Group Comparisons
20 PERMANOVA
Learning Objectives
To explore PERMANOVA, a particularly powerful analytical technique for multivariate data.
To understand the flexibility and limitations of PERMANOVA.
Readings
Anderson (2001)
Key Packages
require(vegan)
Introduction
PERmutational Multivariate ANalysis of VAriance (PERMANOVA) is a permutation-based technique – it makes no distributional assumptions about multivariate normality or homogeneity of variances. It can be thought of visually or geometrically.
PERMANOVA is equivalent to MRPP under certain conditions (Reiss et al. 2009), but is more versatile and widely applicable. Understanding its versatility requires also considering its flexibility and its limitations.
Theory
Recall that ANOVA quantifies the variation within a dataset as the sum of squared differences between points and their mean or centroid. This is used to calculate the total variation (total sum of squares; SST) based on the differences between points and the grand mean. It is also used to calculate the variation within groups (SSW) from the differences between points and their group means. SSW is some subset of SST; variation that is not due to SSW occurs between/among groups (SSB).
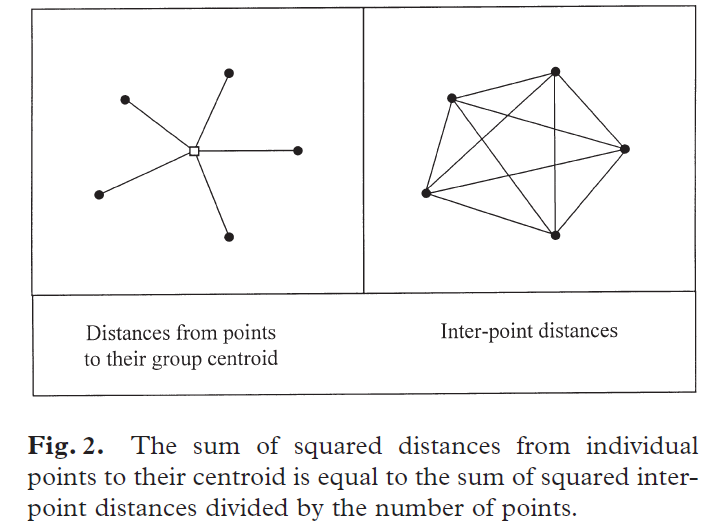
The key insight behind PERMANOVA is that the variation within a group can be calculated directly from a distance matrix. Specifically, the sum of squared differences between points and their centroid is equal to the sum of the squared interpoint distances divided by the number of points. This idea is illustrated below (Fig. 2 from Anderson 2001). It is known as Huygens’ theorem as it was first formulated by Christiaan Huygens – in the 17th century (Anderson et al. 2008)!
Huygens’ theorem means that variation can be calculated without knowing the location of group centroids. Anderson (2015) describes this as geometric partitioning. While this may seem abstract, it is crucial for semimetric distance measures (e.g., Bray-Curtis) that do not satisfy the triangle inequality (please review the Distance Measures chapter if this does not sound familiar). When using a semimetric distance measure, we cannot determine the locations of centroids from the locations of the data. By avoiding this step, basic ANOVA theory can be applied to data summarized with any distance measure. More details, including additional information about the mathematics of PERMANOVA, can be found in Anderson (2001, 2017), McArdle & Anderson (2001), and Anderson et al. (2008).
Knowing that we can calculate variation from the distance matrix means that we can also partition that variation among sources. The resulting technique requires no assumptions about normality and/or homogeneity of variances. The only assumption is that observations are interchangeable under the null hypothesis – the same assumption as with all permutation tests, and the justification for using permutations to assess significance.
The PERMANOVA test statistic, pseudo-F, is modeled directly after the conventional ANOVA F-statistic. It is calculated in the same way, as a ratio of the amount of variation between versus within groups, with the numerator and denominator each weighted by their degrees of freedom. It is 0 or positive, with larger values corresponding to larger proportional importance of the grouping factor. As with other permutational techniques, the distribution of the test statistic is assessed by repeatedly relabelling observational units (i.e., permuting group identities) and recalculating the test statistic.
Since it is based on the distance matrix, PERMANOVA can be applied identically to both univariate and multivariate data. An important, and reassuring, point: the test statistic is identical to the conventional F-statistic when calculated using Euclidean distance for a single variable.
PERMANOVA can be used to analyze complex models, including multiple factors, covariates, and interactions between terms. More information about this is provided in the chapters about complex designs and restricting permutations.
Laughlin et al. (2004) provide an example of how these techniques can be applied to plant community data with repeated measurements (i.e., compositional data from permanent plots sampled in multiple years). Hudson & Henry (2009) used adonis() while Assis et al. (2018) used adonis2(); we discuss both of these functions below. Sharma et al. (2019) use both PERMANOVA (though they don’t specify which function within vegan) and anosim() to examine the microbiome of people and their built environment.
Key Takeaways
PERMANOVA compares the variation between groups to the variation within groups.
The test statistic, pseudo-F, is modeled after the F-statistic from ANOVA. It is 0 or positive, with larger values corresponding to larger proportional importance of the grouping factor.
PERMANOVA is an extremely powerful and flexible technique. It can be applied to data of any dimensionality (including univariate) and expressed through any distance measure. It can also be applied to complex designs, including multiple factors and covariates (continuously distributed predictors).
Basic Procedure
The basic procedure for PERMANOVA is as follows.
- Convert data matrix to a distance matrix, using an appropriate distance measure.
- Square the distance matrix.
- Calculate three partitions of the variation:
- Total variation (total sum of squares; SST) – the sum of squared distances divided by the number of plots.
- Variation within groups (sum of squares within groups; SSW) – calculated identical to SST but separately for each group. Add the value from each group together to yield the overall SSW.
- Variation between groups (sum of squares between groups; SSB) – calculated by simple subtraction (i.e., SSB = SST – SSW).
- Calculate the test statistic. The test statistic is termed a ‘pseudo-F statistic’ to distinguish it from the traditional parametric univariate F-statistic, but is calculated using the same formula:
[latex]PseudoF = \frac{SSB / (t - 1)}{SSW / (N - t)}[/latex]
where SSB and SSW are as calculated above, t is the number of groups, and N is the total number of sample units.
- Assess statistical significance via a permutation test:
- Permute group identities.
- Recalculate pseudo-F statistic
- Repeat the specified number of times. The permutations produce a distribution of pseudo-F values against which the actual pseudo-F statistic value is compared.
- Calculate the P-value as the proportion of permutations that yielded a pseudo-F value equal or greater than the actual data did.
Simple Example, Worked By Hand
Using the data and group identities from our simple example, let’s work through the calculations involved with PERMANOVA.
We begin with our distance matrix. For clarity, the distances between plots from different groups are in bold (view perm.eg$Group to confirm this).
d.eg <- dist(perm.eg[ , c("Resp1", "Resp2")])
| Plot1 | Plot2 | Plot3 | Plot4 | Plot5 | |
| Plot2 | 2.828 | ||||
| Plot3 | 4.123 | 2.236 | |||
| Plot4 | 11.314 | 11.662 | 9.849 | ||
| Plot5 | 9.849 | 9.220 | 7.071 | 4.123 | |
| Plot6 | 12.207 | 12.042 | 10.000 | 2.236 | 3.162 |
Square the distance matrix to yield squared distances among plots.
d.eg_2 <- d.eg^2
| Plot1 | Plot2 | Plot3 | Plot4 | Plot5 | |
| Plot2 | 8 | ||||
| Plot3 | 17 | 5 | |||
| Plot4 | 128 | 136 | 97 | ||
| Plot5 | 97 | 85 | 50 | 17 | |
| Plot6 | 149 | 145 | 100 | 5 | 10 |
Sum the squared distances and divide by the number of plots to obtain the total sum of squares (SST):
[latex]SST = \frac{1049}{6} = 174.83[/latex]
SST <- sum(d.eg_2) / length(perm.eg$Group)
Note that this is Huygens’ theorem in action!
We obviously could combine these steps. For example, here’s a function to combine all of these steps:
SS.calculation <- function(x, variables) {
n <- nrow(x)
x %>%
dplyr::select(any_of(variables)) %>%
dist() %>%
`^`(2) %>%
sum() / n
}
SS.calculation(x = perm.eg,
variables = c("Resp1", "Resp2"))
[1] 174.8333Review the order of operations as outlined in the pipe. One unusual element here is the squaring of the distance matrix; for reasons that I don’t understand I was able to do this using the magrittr pipe (%>%) but not the base R pipe (|>). This function is available in the GitHub repository.
Now let’s calculate the variation within each group (i.e., SSW). We can manually identify the appropriate squared distances from the above matrix and do the calculation:
[latex]SSW = \frac{8 + 17 + 5}{3} + \frac{17 + 5 + 10}{3} = \frac{30}{3} + \frac{32}{3} = 20.67[/latex]
Or, we can use SS.calculation() to calculate the variation within each group – all we need to do is index the object to refer to the group of interest:
SSW.A <- SS.calculation(x = perm.eg |> filter(Group == "A"),
variables = c("Resp1", "Resp2"))
SSW.B <- SS.calculation(x = perm.eg |> filter(Group == "B"),
variables = c("Resp1", "Resp2"))
SSW <- SSW.A + SSW.B
Calculate the variation between groups (SSB) as the difference between SST and SSW:
[latex]SSB = SST - SSW = 174.83 - 20.67 = 154.16[/latex]
SSB <- SST - SSW
Finally, calculate the pseudo-F statistic:
[latex]PseudoF = \frac{SSB / (t - 1)}{SSW / (N - t)} = \frac{154.16 / 1}{20.67 / 4} = 29.83[/latex]
pseudo.F <- (SSB / (2 - 1)) / (SSW / (6 - 2))
Often, we will summarize this information in an ANOVA table:
| Source | df | SS | MS | Pseudo-F |
| Group | 1 | 154.16 | 154.16 | 29.83 |
| Residual | 4 | 20.67 | 5.17 | |
| Total | 5 | 174.83 |
This table contains all of the data that we usually see in an ANOVA output except the P-value. Statistical significance will be determined by permuting the group identities, recalculating the pseudo-F statistic for each permutation, and comparing the observed value against the distribution of values obtained via permutation.
Can you figure out what the pseudo-F statistic is for the permutation of group identities {A,B,A,A,B,B}? As a hint, the squared distance matrix is repeated here, with bold text to indicate distances between plots from different groups.
| Plot1 | Plot2 | Plot3 | Plot4 | Plot5 | |
| Plot2 | 8 | ||||
| Plot3 | 17 | 5 | |||
| Plot4 | 128 | 136 | 97 | ||
| Plot5 | 97 | 85 | 50 | 17 | |
| Plot6 | 149 | 145 | 100 | 5 | 10 |
Implementation in R (vegan::adonis2())
PERMANOVA is becoming increasingly common in community ecology. It is surprising, therefore, that only a few R packages provide it. We are using the main one, vegan.
The LDM (linear decomposition model) package provides another formulation of PERMANOVA. The authors claim that their formulation outperforms those in vegan. It was developed for use with microbiome data and seeks to simultaneously test global hypotheses (i.e., overall assessment of compositional differences among treatments) and individual operational taxonomic units (OTUs; i.e., response variables) (Hu & Satten 2020). Finally, the PERMANOVA package was released in 2021 and updated in late 2022 (Vicente-Gonzalez & Vicente-Villardon 2022). I have not yet evaluated either of these packages.
Several other packages also offer permutational multivariate analysis of variance and appear to be independent, but require the vegan package so often (I’ve not verified all) are using vegan’s functions. These include:
GUniFrac– this package provides Generalized UniFrac distances for comparing microbial communities (Chen et al. 2012). The package description states that it provides three extensions to PERMANOVA: a different permutation scheme (Freedman-Lane), an omnibus test using multiple matrices, and an analytical approach for approximating the PERMANOVA p-value. The PERMANOVA function here isPermanovaG().RVAideMemoire– this package provides miscellaneous functions useful in biostatistics. The PERMANOVA functions here areperm.anova()andadonis.II(). According to the help files:perm.anova()can be applied to 1 to 3 factors. For 2 or 3 factors, the experimental design must be balanced. For 2 factors, the factors can be crossed with or without interaction, or nested. The second factor can be a blocking (random) factor. For 3 factors, design is restricted to 2 fixed factors crossed (with or without interaction) inside blocks (third factor).adonis.II()is a wrapper tovegan::adonis()but performs type II tests (whereasvegan::adonis()performs type I tests).
biodiversityR– this package provides a graphical user interface (GUI) for community analysis – this can be useful in the moment but is difficult to script. Its website includes a downloadable book (Kindt & Coe 2005) about the analysis of community data, focusing specifically on trees. The PERMANOVA function here isnested.npmanova().- ade4::amova?
- Capscale; anova.cca of dbrda
- The
microbiomeSeqpackage combines PERMANOVA with an ordination, so the statistical test is automatically accompanied by a visualization of the data. However, its website (http://userweb.eng.gla.ac.uk/umer.ijaz/projects/microbiomeSeq_Tutorial.html) is from 2017 yet has a disclaimer that it is still in development. I haven’t explored the function it uses to conduct the PERMANOVA. smartsnp–smart_permanova()is based onvegan::adonis().
PERMANOVA can be conducted using the adonis2() and adonis() functions in vegan. These two functions differ in scope, speed – the help file notes that adonis2() can be much slower than adonis() – and output. Confusingly, these functions yield objects of different classes. As of version 2.6-4, the vegan maintainers note that adonis() will be eliminated soon, so I do not describe it below. Also, some of the arguments described below for adonis2() are not available in older versions of this package.
vegan::adonis2()
As indicated by its name, adonis2() is a replacement for adonis(). Its usage is:
adonis2(
formula,
data,
permutations = 999,
method = "bray",
sqrt.dist = FALSE,
add = FALSE,
by = "terms",
parallel = getOption("mc.cores"),
na.action = na.fail,
strata = NULL,
...
)
The main arguments are:
formula– model formula such asY ~ A + B * C. See the ‘Complex Models‘ chapter for more details of how to specify a model formula. For now, the key point is that we are specifying that we want to analyze the response (Y) as a function of (~) one or more explanatory variables (A,B,C). The class ofYdetermines what is done:- If
Yis a dissimilarity matrix (i.e., output fromdist()orvegdist()), the pseudo-F statistic is calculated directly. - If
Yis a data frame or matrix containing data,vegdist()is applied to calculate the distance matrix first and then the pseudo-F statistic is calculated.
- If
data– the data frame in which the explanatory variables are located. Sample units are assumed to be in the same order indataas in the rows ofY.permutations– number of permutations to conduct to assess the significance of the pseudo-F statistic, or a list of permutation instructions obtained using thehow()function. This latter option is particularly important for PERMANOVA, as it is necessary when permutations need to be restricted – see that chapter for details. Default is to conduct 999 permutations without any restrictions to the permutations.method– method of calculating the distance matrix, if needed (i.e., ifYin the formula is not a distance matrix). Thevegdist()function is used to do these calculations, which is why the default is Bray-Curtis, though you can specify any distance method available invegdist().sqrt.dist– take square root of dissimilarities. This is one way to make semimetric dissimilarities behave in a more ‘Euclidean’ manner. Default is to not ‘euclidify’ them in this fashion (FALSE).add– add a constant to the dissimilarities so that no eigenvalues are negative. There are two options here, which we’ll discuss in the context of Principal Coordinates Analysis (PCoA). Default is to not add this value (FALSE).by– How to deal with other terms when assessing significance. See below for additional discussion of types of sums of squares. Four options are available:terms– tests terms in sequential order, from first to last in formula. The default. This is the Type I or ‘sequential’ sums of squares that you may recall from basic statistics courses.margin– tests the marginal effect of each term, or its effect after accounting for all other terms in the model. This is Type III or ‘partial’ sums of squares that you may recall from basic statistics courses. If applied to a model with an interaction term, this will return just the interaction term and ignore the main effects.onedf– tests terms as a series of contrasts, each with one df.NULL– tests all terms together rather than individually.
parallel– option for parallel processingna.action– what to do if explanatory variables are missing. Default is to not continue (na.fail).strata– groups that permutations are to be restricted within. Can alternatively be specified through thepermutations()function, which is discussed in this chapter about restricted permutations.
The results from adonis2() are saved to an object of class “anova.cca“. This object is also recognized as belonging to classes “anova” and “data.frame“. It currently includes the degrees of freedom (Df), sums of squares (SumOfSqs), partial R2 (R2), pseudo-F statistic (F), and P-value for each term (Pr(>F)) along with the pseudo-F statistics from the permutations (attr(x, "F.perm")) and lots of details about how permutations were conducted.
The partial R2 for each term is calculated as the SS for the term as a proportion of SST. The partial R2 values may NOT sum to 1 if you are testing multiple factors using a marginal model (by = "margin") – see the discussion of types of sums of squares in the ‘Complex Models‘ chapter.
Simple Example
simple.result.adonis2 <- adonis2(
perm.eg[ , c("Resp1", "Resp2")] ~ Group,
data = perm.eg,
method = "euc"
)
Calling this object prints the ANOVA table in the console.
Permutation test for adonis under reduced model
Terms added sequentially (first to last)
Permutation: free
Number of permutations: 719
adonis2(formula = perm.eg[, c("Resp1", "Resp2")] ~ Group, data = perm.eg,
method = "euc")
Df SumOfSqs R2 F Pr(>F)
Group 1 154.167 0.88179 29.839 0.1
Residual 4 20.667 0.11821
Total 5 174.833 1.00000
Verify the calculations we made by hand above. The ANOVA table is identical to what we calculated, though the order of the information is slightly different. Also, this table includes two elements that we did not calculate:
- The partial R2 value for each term. As noted above, this is calculated as the SS for that term as a proportion of the total SS for the dataset. For example, the partial R2 for Group = 154.167 / 174.833 = 0.88179
- A permutation-based P-value for each term being tested (i.e., other than Residual and Total). Although Group has a large pseudo-F value, it is not statistically significant. This is a reflection of the limited number of permutations for a small sample size.
Grazing Example
grazing.result.adonis2 <- adonis2(
Oak1 ~ grazing,
method = "bray"
)
Permutation test for adonis under reduced model
Terms added sequentially (first to last)
Permutation: free
Number of permutations: 999
adonis2(formula = Oak1 ~ grazing, method = "bray")
Df SumOfSqs R2 F Pr(>F)
grazing 1 0.6491 0.05598 2.6684 0.001 ***
Residual 45 10.9458 0.94402
Total 46 11.5949 1.00000
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Grazing clearly affects community composition, though it accounts for a relatively small amount of the variation in composition (5.6%).
We did not specify the data argument because grazing was saved as a separate object. If we wanted to, we could have called it by its column name within Oak (or Oak_explan). Also, if we had specified the distance matrix instead of the data matrix for Y, the method argument would have been unnecessary:
adonis2(Oak1.dist ~ grazing)
Extensions of PERMANOVA
I’ve introduced PERMANOVA as a means of comparing groups, but it is more broadly described as a linear model. Recall that ANOVA is a specific instance of a linear model in which the explanatory variable is categorical. Linear models can be used to compare groups (categorical variables) and to regress one continuously distributed variable on another. As such, PERMANOVA can be used to analyze any linear model. This is sometimes referred to more generally as a ‘distance-based linear model’ (DISTLM) or as distance-based redundancy analysis (dbRDA) (Legendre & Anderson 1999; McArdle & Anderson 2001).
The adonis2() function can handle both ANOVA and regression models. Each variable is treated as categorical or continuous based on its class. Therefore, it is helpful to always check that the degrees of freedom in a model output correspond to what you expected. A variable that is categorical will have 1 fewer df than it has levels, whereas a variable that is continuous will only have 1 df. More information is available in the ‘Complex Models‘ chapter.
Limitations of vegan::adonis2()
Although PERMANOVA is an extremely powerful technique, its implementation in R has some limitations. Topics like the choice of sums of squares mentioned above are not unique to adonis2(). Two other issues to be aware of are how to handle random effects and how to decide what to permute.
Random Effects
Currently, adonis2() does not permit the specification of random effects. An alternative way to account for random effects is to use sequential sums of squares and fit the term that would otherwise be designated as a random effect as the very first term in the model. Doing so results in a model in which as much variation as possible is explained with that term before the other factor(s) are evaluated. However, using this approach is ‘expensive’ in terms of df.
For example, if plots were arranged in blocks (e.g., the whole plots of a split-plot design), block ID could be included as the first term in the model so that the variation among blocks is accounted for before testing other terms. Permutations would also have to be restricted to reflect this design.
Choice of What to Permute
In our earlier examples of permutations, we permuted the group assignment of each sample unit. We could alternatively have kept the group assignments unchanged but permuted the rows of the response matrix. Furthermore, the raw data can be permuted or the residuals obtained from a model can be permuted. In adonis2(), we are not able to specify which of these methods to use. However, this is likely a relatively minor issue: Anderson et al. (2008) note that these methods give very similar results.
Non-R Formulations of PERMANOVA
If the limitations of adonis2() are problematic for your analysis, you may want to explore a non-R formulation of PERMANOVA.
The PERMANOVA+ extension to PRIMER (http://www.primer-e.com/) is an extremely powerful and versatile way to use PERMANOVA. It was coded by Dr. Anderson (author of the 2001 article that developed this technique and introduced it to ecological audiences) and includes an extensive help manual (Anderson et al. 2008). Among other features, it permits you to choose which type of SS you would like to use and to analyze unbalanced data, random effects, etc. The basic approach requires that you create a design file containing the necessary information for partitioning the data based on the factors and experimental design, and then run the PERMANOVA on the distance matrix, choosing that design file during the analysis.
In PC-ORD (https://www.pcord.com/), PERMANOVA can be used to analyze fairly simple designs. Factors are identified in a second matrix (i.e., where explanatory variables are identified). Any of the distance measures available through PC-ORD can be used. Pairwise comparisons can be made to evaluate which groups differ in factors with more than two levels. However, the formulation of PERMANOVA in PC-ORD (version 6) has a number of limitations:
- Requires balanced data (same number of sample units in each group)
- Limited to one or two factors
Tang et al. (2016) presented PERMANOVA-S, an extension of PERMANOVA for microbiome data that i) can account for ‘confounders’ (confounding variables that are correlated with both the response matrix and explanatory variables), and ii) can compare conclusions across multiple distance measures simultaneously. For example, an analysis could compare the conclusions if based on Bray-Curtis distances (weighted by abundance) or on Jaccard distance (based on presence/absence data). A free program, in the C language, used to be available through Tang’s website (https://tangzheng1.github.io/tanglab/software.html) though as of January 2024 it no longer appears there.
Conclusions
PERMANOVA is an extremely powerful and versatile technique. It has strong parallels with ANOVA and thus is familiar to most ecologists.
The flexibility of PERMANOVA allows it to be used in complex models, but this means it can also be used incorrectly. See additional ideas in the chapters about complex models, controlling permutations, and restricting permutations.
References
Anderson, M.J. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecology 26:32-46.
Anderson, M.J. 2015. Workshop on multivariate analysis of complex experimental designs using the PERMANOVA+ add-on to PRIMER v7. PRIMER-E Ltd, Plymouth Marine Laboratory, Plymouth, UK.
Anderson, M.J. 2017. Permutational Multivariate Analysis of Variance (PERMANOVA). Wiley StatsRef: Statistics Reference Online. DOI: 10.1002/9781118445112.stat07841
Anderson, M.J., and C.J.F. ter Braak. 2003. Permutation tests for multi-factorial analysis of variance. Journal of Statistical Computation and Simulation 73:85-113.
Anderson, M.J., R.N. Gorley, and K.R. Clarke. 2008. PERMANOVA+ for PRIMER: guide to software and statistical methods. PRIMER-E Ltd, Plymouth Marine Laboratory, Plymouth, UK. 214 p.
Assis, D.S., I.A. Dos Santos, F.N. Ramos, K.E. Barrios-Rojas, J.D. Majer, and E.F. Vilela. 2018. Agricultural matrices affect ground ant assemblage composition inside forest fragments. PLoS One 13(5):e0197697.
Chen, J., K. Bittinger, E.S. Charlson, C. Hoffmann, J. Lewis, G.D. Wu, R.G. Collman, F.D. Bushman, and H. Li. 2012. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 28:2106-2113.
Hu, Y-J., and G.A. Satten. 2020. Testing hypotheses about the microbiome using the linear decomposition model (LDM). Bioinformatics 36:4106-4115.
Hudson, J.M.G., and G.H.R. Henry. 2009. Increased plant biomass in a high Arctic heath community from 1981 to 2008. Ecology 90:2657-2663.
Kindt, R., and R. Coe. 2005. Tree diversity analysis: a manual and software for common statistical methods for ecological and biodiversity studies. World Agroforestry Centre (ICRAF), Nairobi, Kenya. http://apps.worldagroforestry.org/downloads/Publications/PDFS/b13695.pdf
Laughlin, D.C., J.D. Bakker, M.T. Stoddard, M.L. Daniels, J.D. Springer, C.N. Gildar, A.M. Green, and W.W. Covington. 2004. Toward reference conditions: wildfire effects on flora in an old-growth ponderosa pine forest. Forest Ecology and Management 199:137-152.
Manly, B.F.J., and J.A. Navarro Alberto. 2017. Multivariate statistical methods: a primer. Fourth edition. CRC Press, Boca Raton, FL.
McArdle, B.H., and M.J. Anderson. 2001. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82:290-297.
Sharma, A., M. Richardson, L. Cralle, C.E. Stamper, J.P. Maestre, K.A. Stearns-Yoder, T.T. Postolache, K.L. Bates, K.A. Kinney, L.A. Brenner, C.A. Lowry, J.A. Gilbert, and A.J. Hoisington. 2019. Longitudinal homogenization of the microbiome between both occupants and the built environment in a cohort of United States Air Force cadets. Microbiome 7:70.
Tang, Z-Z., G. Chen, and A.V. Alekseyenko. 2016. PERMANOVA-S: association test for microbial community composition that accommodates confounders and multiple distances. Bioinformatics 32:2618-2625.
Vicente-Gonzalez, L., and J.L. Vicente-Villardon. 2022. PERMANOVA: Multivariate analysis of variance based on distances and permutations. R Package, v.0.2.0. https://cran.r-project.org/package=PERMANOVA
Media Attributions
- Anderson.2001_Figure2