Follow-Up Tests
46 ISA
Learning Objectives
To understand the calculations behind ISA:
- Classical approach based on abundance data
- Considerations of combinations of groups
- Simplified approach based on presence/absence data
To summarize and apply the results of an ISA.
To consider additional ways that ISA could be used.
Readings (recommended)
Dufrêne & Legendre (1997)
Key Packages
require(vegan, indicspecies)
Introduction
Indicator Species Analysis (ISA) was originally developed by Dufrêne & Legendre (1997) as a technique to identify species that are strongly associated with a particular group. One motivation for developing this technique was to provide a tool for determining how many groups to focus on in a cluster analysis.
ISA has been extended in several ways since it was originally described:
- De Cáceres & Legendre (2009) showed how to use ISA to identify species that are associated with multiple groups.
- I (Bakker 2008) demonstrated that ISA produces very similar results if applied to presence/absence data rather than abundance data. Implementation is identical, but with a matrix of presence/absence data instead of abundance data.
- In the same paper, I also showed how ISAs from different sites can be combined. I will not explore this idea further here, but it is available through
indicspecies::signassoc(). - Baker & King (2010) developed TITAN to identify species that are associated with high or low values of a continuously distributed variable.
I will explain the way this was first proposed (i.e., the classical ISA) and then the extensions to multiple groups and to presence/absence data. I consider TITAN separately.
The Classical ISA
ISA is a remarkably simple technique that I think is underappreciated. Advantages include:
- easy to calculate
- easy to interpret
- statistical significance is assessed via permutation tests
- can be used with any categorical classification (a priori or a posteriori)
- is calculated independently for each species
ISA requires a typology, or classification of sample units into groups. As noted below, multiple groupings can be tested, and these groupings can be hierarchical or non-hierarchical.
ISA involves calculating the specificity (Aij; relative abundance) and fidelity (Bij; relative frequency) of species i in group j. These values are then multiplied together to yield the test statistic, the Indicator Value (IVij).
| Formula | Verbal Interpretation | Range | |
| Specificity / Relative Abundance: | [latex]A_{ij} = \frac {\bar{x}_{ij}}{\sum_{j} \bar{x}_{i\bullet}}[/latex] | Mean cover of species i in group j as a proportion of its mean cover in all groups | 0 to 1 |
| Fidelity / Relative Frequency: | [latex]B_{ij} = \frac {n_{ij}}{n_{\bullet j}}[/latex] | Proportion of plots in group j on which species i occurs | 0 to 1 |
| Indicator Value: | [latex]IV_{ij} = A_{ij} \times B_{ij} \times 100[/latex] | As proposed by Dufrêne & Legendre (1997) | 0 to 100 |
| [latex]IV_{ij} = \sqrt {A_{ij} \times B_{ij}}[/latex] | As reported in indicspecies::multipatt() |
0 to 1 |
where
- [latex]\bar{x}_{ij}[/latex] is the mean cover of species i within group j
- [latex]\sum_{j} \bar{x}_{i\bullet}[/latex] is the sum of the mean cover of species i in all groups
- [latex]n_{ij}[/latex] is the number of plots in group j occupied by species i
- [latex]n_{\bullet j}[/latex] is the total number of plots in group j.
The key statistic is the Indicator Value (IndVal or IV or IVij). Some notes about its interpretation:
- IVij is 0 when species i is absent from group j, and is at its maximum (1 or 100) when species i occurs on all plots within group j and does not occur in other groups.
- The highest IVij values will occur for species that are common in one group and absent from other groups within the typology.
- Uncommon species will have low Bij values and therefore low IVij values.
- Ubiquitous species (i.e., species present in all groups at a given level of the typology) have high Bij values and therefore high IVij values.
- Dufrêne & Legendre (1997) suggest that species i be considered a ‘strong’ indicator of group j if IVij > 25. There are multiple ways to arrive at this value. For example:
- If a species is present on at least 50% of the plots in group j (i.e., Aij = 0.5) and also has at least 50% of its total abundance in group j (i.e., Bij = 0.5)
- If a species is present on at least 25% of the plots in group j (i.e., Aij = 0.25) and also has all of its total abundance in group j (i.e., Bij = 1.0)
- If a species is present on all pots in group j (i.e., Aij = 1.0) and also has at least 25% of its total abundance in group j (i.e., Bij = 0.25)
- The minimum of IVij = 25 for a strong indicator as proposed by Dufrêne & Legendre (1997) is equivalent to IVij = 0.5 in the formulation reported by
indicspecies::multipatt()which takes the square root of this value. - The figure below illustrates isoclines of IV:
- Left: product of Aij and Bij (though expressed as a proportion rather than a percentage), as proposed by Dufrêne & Legendre (1997)
- Right: squareroot of product of Aij and Bij, as reported by
indicspecies::multipatt()

Species i is tentatively considered an indicator of the group j in which its IV is largest. The statistical significance of these values is assessed using permutation tests. These tests are necessary because ubiquitous species have high IVij values but are not helpful in distinguishing one group from another.
Indicators of Combinations of Groups
As originally proposed, ISA only considers species as potential indicators of one group or another. For example, imagine that you had obtained samples from forest, grassland, and river. The classical ISA identifies species that are indicators of forest OR grassland OR river. However, a species that occurred in both forest AND grassland would not be identified as an indicator of either habitat.
Miquel De Cáceres and colleagues (De Cáceres & Legendre 2009; De Cáceres et al. 2010) extended the classical ISA to enable the identification of indicators of combinations of groups. They argued that this extension provides a way to identify species with differing niche breadths (i.e., niches spanning more than one category).
For example, in the habitat example that I just described, the method proposed by De Cáceres et al. tests for indicators of individual habitats (forest OR grassland OR river) and for indicators of combinations of habitats (e.g., (forest + grassland) OR river). The restcomb argument within indicspecies::multipatt() provides control over which combinations are considered.
Simplified ISA: Presence/Absence Data
Dufrêne & Legendre (1997) also proposed a simplified ISA based on presence/absence data. For the simplified ISA, the formula for specificity (Aij) is modified to:
[latex]A_{ij} = \frac {n_{ij}}{n_{i\bullet}}[/latex]
where
- [latex]n_{ij}[/latex] is the number of plots in group j occupied by species i
- [latex]n_{i\bullet}[/latex] is the total number of plots occupied by species i
Verbally, specificity is now the proportion of plots occupied by species i that are located in group j. Bij and IVij are calculated as above.
In practice, a simplified ISA can be implemented by transforming the data to binary data (i.e., 0 = absence, 1 = presence) and calculating the ISA on the transformed data. If a species is equally abundant on all plots where it occurs, the classical and simplified ISAs yield identical results. As variability in abundance among plots increases, the classical and simplified ISAs give differing weights to Aij, and therefore yield different results.
A potentially large advantage of the simplified ISA is that the data are easily summarized and displayed. To summarize the data necessary for the classical ISA, you need the raw data (i.e., abundance of species i on every plot j). In contrast, the data needed for the simplified ISA can be succinctly summarized. Referring to the formulae above, you can see that only three values are required to calculate the simplified ISA for a given species:
- [latex]n_{ij}[/latex] = the number of plots in group j occupied by species i
- [latex]n_{i\bullet}[/latex] = the total number of plots occupied by species i
- [latex]n_{\bullet j}[/latex] = the total number of plots in group j
The simplified ISA can be conducted using the classical approach (each group separately) or the extension to combinations of groups.
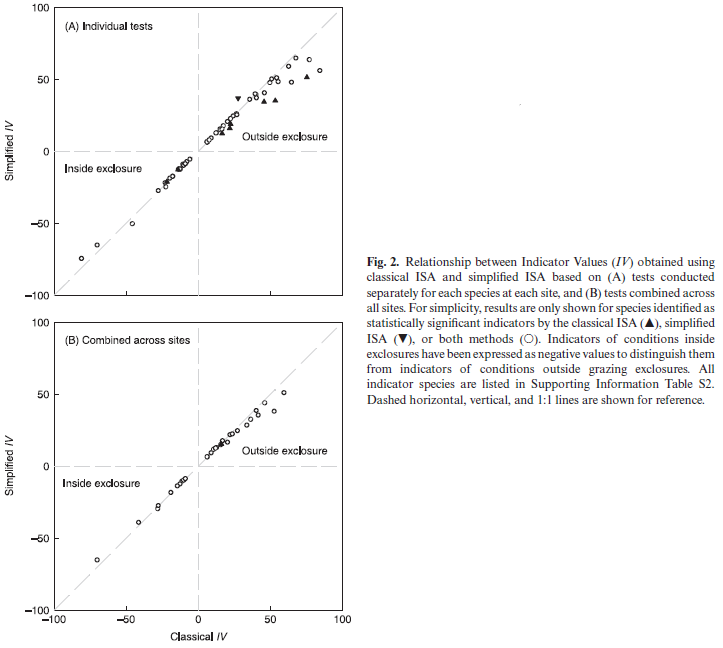
In an earlier study (Bakker 2008), I used classical (abundance-based) and simplified (presence/absence-based) ISAs to identify indicators of grazing treatment (Figure 2; reproduced below). There were 206 unique tests, each consisting of one species at one site. Summary conclusions:
- In 94% (193 of 206) of tests, the classical and simplified ISAs identified the same grazing treatment as having the maximum IV.
- In no test did the two methods identify opposite grazing treatments as statistically significant indicators.
- Overall, 62 species were identified as statistically significant indicators of grazing treatment. Of these:
- 85% (53 of 62) were identified by both methods
- 13% (8 of 62) were identified by the classical ISA only
- 2% (1 of 62) were identified by the simplified ISA only
Published ISA Examples
Zald et al. (2020) identified plant species associated with different seasons and intervals of burning.
Pillsbury et al. (2019) used ISA to identify diatoms associated with high and low nitrogen and phosphorus conditions.
Martin & Hamman (2016) used ISA to identify the fire severity class associated with each of several plant life forms.
Severns & Sykes (2020) propose that phytopathologists use ISA to identify the fungi associated with plant diseases.
ISA in R (indicspecies::multipatt())
There are several functions that can conduct ISA in R. They vary in some details, such as how IV is reported. We will focus on indicspecies::multipatt() but you are welcome to explore others such as labdsv::duleg() and adiv::dbMANOVAspecies() (Ricotta et al. 2021) if interested.
The usage of multipatt() is:
multipatt(x,
cluster,
func = "IndVal.g",
duleg = FALSE,
restcomb = NULL,
min.order = 1,
max.order = NULL,
control = how(),
permutations = NULL,
print.perm = FALSE
)
The key arguments are:
x– the plot x species data tablecluster– a vector consisting of the group to which each plot belongs. Although this is called ‘cluster’, the groups do not have to come from a cluster analysis.func– function to calculate the strength of the association of each species with the groups. Options are"IndVal","IndVal.g","r", and"r.g".IndValandIndVal.gare the IV mentioned above, whilerandr.gare correlations. The ‘.g’ suffix indicates that they include a correction for sample sizes, and are the recommended functions to use.IndVal.gis the default.duleg– whether to consider groups of sites (FALSE; default) or not (TRUE). Argument name is an abbreviation of Dufrêne & Legendre (1997), who only considered indicators of individual sites. Ifduleg = TRUE, this conducts the classical ISA that they originally proposed.restcomb– a vector of integers that restricts attention to groups of sites that make ecological sense to the analyst. Not applicable if doing a classical ISA.min.order– minimum number of sites (i.e., levels of a factor) to include in groups of sites. Default is 1 (i.e., individual sites can be their own groups).max.order– maximum number of sites (i.e., levels of a factor) to include in groups of sites. For example, ifmax.order = 2, then two levels can be combined into a group but a group including three levels will not be tested.control– options for controlling how permutations are conducted. This uses the samepermute::how()function that we saw in our group comparison methods.
The resulting object is of class ‘multipatt’ with several elements, including:
comb– a plot x combination matrix, with a column for every combination of groups tested. The values within this matrix are 1 if that plot is included in that combination of groups and 0 otherwise. The column number is the integer value by which these combinations are referenced in other elements.A– a species x combination matrix, with a column for every combination of groups tested. The values are the specificity component of IV for that species in that combination of groups.B– a species x combination matrix, with a column for every combination of groups tested. The values are the fidelity component of IV for that species in that combination of groupssign– a data table summarizing the results for each species:- The ‘
index’ column contains an integer denoting the combination of groups in which the species had its maximum IV. - The ‘
stat’ column reports the square root of the IV. - The ‘
p.value’ column reports the permutation-based p-value associated withstat. Note that there is no p-value when the maximum IV is in the largest combination of groups (i.e., all plots in a single group) as there are no permutations possible.
- The ‘
Applying summary() to the resulting object focuses on those species that meet desired criteria. The default is to show those species that are statistically significant at alpha = 0.05. See ?summary.multipatt for details of other criteria.
One caution about only viewing statistically significant taxa is that the p-value is permutation-based and therefore can vary; so species may be identified as significant indicators in one run but not in another run. Options to minimize this issue are to use a set group of permutations (set.seed()) and/or to conduct a large number of permutations.
Oak Example: Indicators of Grazing Treatments
Let’s see which species are strongly associated with one grazing status over the others. We will do this using the three approaches described above: classical ISA, ISA with multiple groups, and simplified ISA (also with multiple groups).
To ensure that results are directly comparable amongst analyses, we will use the same set of permutations in each analysis.
Classical ISA
To use the classical ISA, we specify x and cluster, and include the argument ‘duleg = TRUE’:
library(indicspecies)
set.seed(42)
Graz.ISA <- multipatt(x = Oak1,
cluster = Oak_explan$Grazing,
duleg = TRUE)
summary(Graz.ISA)
Multilevel pattern analysis
---------------------------
Association function: IndVal.g
Significance level (alpha): 0.05
Total number of species: 103
Selected number of species: 6
Number of species associated to 1 group: 6
Number of species associated to 2 groups: 0
List of species associated to each combination:
Group Always #sps. 2
stat p.value
Popr 0.74 0.04 *
Agte 0.72 0.03 *
Group Never #sps. 2
stat p.value
Prav.s 0.834 0.005 **
Syal.s 0.757 0.005 **
Group Past #sps. 2
stat p.value
Rhdi.s 0.690 0.015 *
Aqfo 0.544 0.035 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Six of the 103 species are identified as significant indicators. All such indicators are associated with a single group (as designed in the classical ISA). In this case:
- Popr and Agte are indicators of always grazed stands
- Prav.s and Syal.s are indicators of never grazed stands
- Rhdi.s and Agfo are indicators of stands that were grazed only in the past
Note that it is simply chance that there are equal numbers of indicators for each grazing treatment. There could easily be uneven numbers of indicators, and groups for which no indicators are identified.
Each species is identified by the unique code that named its column. We can use these codes to look their names in our Oak_spp object and then draw on our ecological knowledge of the system to interpret them.
ISA with Multiple Groups
Our grazing example includes three levels that can be combined into seven groups:
| Group | Possible Ecological Interpretation |
| Always | Continually grazed |
| Never | Never grazed |
| Past | No longer grazed |
| Never+Past | Not currently grazed |
| Always+Never | No change in grazing practice? |
| Always+Past | Grazed in the past, regardless of current grazing practices |
| Always+Never+Past | All plots |
(these combinations of groups are identified in Graz.ISA.2$comb below)
The multipatt() function automatically combines the levels of our grouping variable into groups of levels. If desired, we can use the restcomb argument to restrict our attention to particular combinations of groups. For example, we might decide that the group ‘Always+Never’ is ecologically questionable and therefore exclude it from consideration (I have not done so here, however).
ISA with multiple groups is the default in multipatt():
set.seed(42)
Graz.ISA.2 <- multipatt(x = Oak1,
cluster = Oak_explan$Grazing)
summary(Graz.ISA.2)
Multilevel pattern analysis
---------------------------
Association function: IndVal.g
Significance level (alpha): 0.05
Total number of species: 103
Selected number of species: 9
Number of species associated to 1 group: 1
Number of species associated to 2 groups: 8
List of species associated to each combination:
Group Past #sps. 1
stat p.value
Aqfo 0.544 0.035 *
Group Always+Past #sps. 6
stat p.value
Sado 0.841 0.005 **
Popr 0.808 0.010 **
Elgl 0.786 0.030 *
Hola 0.753 0.035 *
Agte 0.727 0.045 *
Acmi 0.606 0.015 *
Group Never+Past #sps. 2
stat p.value
Prav.s 0.873 0.015 *
Ronu.s 0.705 0.045 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 In this analysis, 9 of the 103 species are identified as indicators. Some indicators are associated with 1 group and some are associated with 2 groups. In this case there are:
- One indicator (Agfo) of stands that were only grazed in the past
- Six indicators (Sado, Popr, Elgl, Hola, Agte, and Acmi) of stands that were either always grazed or only grazed in the past (i.e., not never grazed)
- Two indicators (Prav.s, Ronu.s) of stands that were not currently grazed (i.e., either Never or Past grazed)
Allowing for indicators to be identified across multiple groups has changed some of the indicators. For example, there are no indicators of stands that were only Never grazed whereas in the above analysis two species were indicators of this group. Aqfo is still identified as an indicator of a single group (Past), so its test statistic is identical here to what was calculated with the Classical ISA.
Simplified ISA (Presence/Absence Data)
To analyze these data using presence/absence data, we need to relativize the data in that way first. We’ll use the multi-group ISA here.
Oak1.pres <- decostand(Oak1, "pa")
set.seed(42)
Graz.ISA.pres <- multipatt(x = Oak1.pres,
cluster = Oak_explan$Grazing)
summary(Graz.ISA.pres)
Multilevel pattern analysis
---------------------------
Association function: IndVal.g
Significance level (alpha): 0.05
Total number of species: 103
Selected number of species: 8
Number of species associated to 1 group: 2
Number of species associated to 2 groups: 6
List of species associated to each combination:
Group Past #sps. 2
stat p.value
Frla.s 0.565 0.050 *
Aqfo 0.544 0.035 *
Group Always+Past #sps. 6
stat p.value
Sado 0.862 0.005 **
Brla 0.844 0.005 **
Elgl 0.788 0.020 *
Popr 0.771 0.005 **
Acmi 0.609 0.010 **
Cyec 0.511 0.030 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Eight of the 103 species are identified as indicators, and some indicators are associated with 1 group and some are associated with 2 groups. In this case there are:
- Two indicators (Frla.s, Agfo) of stands that were only grazed in the past
- Six indicators (Sado, Brla, Elgl, Popr, Acmi, and Cyec) of stands that were either always grazed or only grazed in the past (i.e., not never grazed)
Results based on presence/absence data are broadly similar to, but not identical, to those above where we also incorporated species abundance information.
Summarizing ISA Results
An interesting element is to compare analyses and identify which indicators have changed in status. This can be done on the basis of the magnitude and significance of IV. Here, we’ll just focus on those species that were identified as significant indicators and compare these across the three ISAs that we conducted. I’ve sorted this list alphabetically by species code.
| Species | Classical ISA | Multiple Group ISA | Simplified ISA |
| Acmi | Always+Past | Always+Past | |
| Agte | Always | Always+Past | |
| Aqfo | Past | Past | Past |
| Brla | Always+Past | ||
| Cyec | Always+Past | ||
| Elgl | Always+Past | Always+Past | |
| Frla.s | Past | ||
| Hola | Always+Past | ||
| Popr | Always | Always+Past | Always+Past |
| Prav.s | Never | Never+Past | |
| Rhdi.s | Past | ||
| Ronu.s | Never+Past | ||
| Sado | Always+Past | Always+Past | |
| Syal.s | Never |
Some observations:
- Aqfo was a consistent indicator of the same group (Past) regardless of how the analysis was conducted.
- Several species that were identified as indicators of a single group in the classical ISA were determined to be even stronger indicators of a combination of groups in the multiple group and/or simplified ISAs.
- Several species were not identified as indicators of a single group in the classical ISA but were identified as strong indicators of a combination of groups in the other analyses.
- Species that were identified as significant indicators in the multiple group ISA but not in the simplified ISA differ more strongly in abundance among groups.
- None of the shrubs (species codes ending in ‘.s’) were indicators of stands that were currently grazed (Always), perhaps reflecting their sensitivity to livestock.
- Indicators of the ‘Never’ and ‘Always’ groups were only detected in the Classical ISA; most of these species were more strongly associated with a combination of groups in the other analyses.
- The complete results for each analysis includes all species regardless of their statistical significance – we could look at this to see which combination they were most strongly associated with.
Additional Applications of ISA
Hierarchical Analyses
Dufrêne & Legendre’s (1997) original description of ISA was as a method to identify how many groups to focus on in a hierarchical cluster analysis. For example, they identified the number of groups in the cluster analysis at which each species achieved its largest IV, and then tallied those up – the height in the dendrogram that produced the greatest number of largest IVs was adopted as the solution and those groups were chosen.
The hierarchical structure of a dendrogram can be easily incorporated into the multi-group ISA (restcomb argument), combining groups in the order that they are fused in the dendrogram, and omitting those that are not supported by the dendrogram.
Non-Hierarchical Analyses
Not all data have a hierarchical typology. For example, if you were interested in two explanatory variables, you could conduct an ISA for each of them and then combine the results.
In one project, I was interested in indicators of overstory condition (either in the open or under tree canopy) and of grazing treatment (inside or outside livestock exclosures). I did an ISA of each of these, and found that some species were indicators of overstory condition, some of grazing treatment, some of a combination of the two, and some of neither:
| Cross-classification of species based on indicator status based on grazing treatment (rows) and overstory condition (columns). Unpublished data, J.D. Bakker. | |||
| In Open | Neither | Under Canopy | |
| Outside | Achillea millefolium | Elymus elymoides | Trifolium longipes |
| Neither | Sporobolus interruptus | Bromus tectorum | |
| Inside | Hesperostipa comata | Muhlenbergia montana | Festuca arizonica |
These comparisons are more ecologically nuanced than just thinking about a single factor. For example, it reflects the fact that different factors shape the distributions of different species.
Consistency of Indicators Among Sites
One aspect of ISA that hasn’t received much attention to date is the consistency of indicators across replicate sites or studies. Meta-analytical techniques can be used to combine the ISA results from multiple sites and thereby assess the consistency of indicators (see Bakker 2008 for details).
For example, areas inside and outside a grazing exclosure at each of five sites in northern Arizona were sampled in 1941. I used these data to examine whether Muhlenbergia montana, a common grass, was an indicator of conditions inside or outside livestock exclosures. To do so, I analyzed each site separately and then combined the results into an overall value for each grazing treatment:
| Inside | Outside | ||||
| Site | IV | P-value | IV | P-value | |
| Big Fill | 36.1 | 0.5882 | 38.3 | 0.4060 | |
| Black Springs | 46.0 | 0.0004 | 3.4 | 1.0000 | |
| Fry Park | 92.1 | 0.0001 | 0.0 | 1.0000 | |
| Reese Tank | 59.2 | 0.0563 | 40.8 | 0.9450 | |
| Rogers Lake | 53.1 | 0.0001 | 0.5 | 1.0000 | |
| Overall | 59.2 | 0.0001 | 15.5 | 1.0000 | |
Although M. montana was clearly not a statistically significant indicator of conditions inside exclosures at one site, and was marginally significant at a second site, it was significant at three other sites. When considered together, it was clearly an indicator of conditions inside exclosures overall. I did similar analyses with a number of other species and found that species that occurred on multiple sites were more likely to be indicators than those that occurred on a single site.
Meta-Analyses
Since ISA is calculated separately for each species, the data can also be summarized separately for each species – for example, there is no need to keep track of which plot each species occurred in. This is particularly useful for the simplified ISA, which only requires three values as noted above.
If the relevant data are reported in the literature, they can be used to assess the generality of indicator species using independent datasets. Furthermore, since ISAs are conducted independently for each species, studies can be drawn from across the distributional range of each species (Bakker 2008).
Species Combinations
De Cáceres et al. (2012) proposed that ISA could also be used to identify combinations of species that distinguish sites. This might occur, for example, if two species are functionally equivalent or even if a taxon was identified to the species level in some plots but to the genus level in other plots (e.g., Poa pratensis, Poa spp.).
This approach can be implemented using indicspecies::combinespecies() to create combinations of species, up to some max.order. See De Cáceres (2023) for details.
Recommendations for ISA
Some recommendations for using ISA, particularly with respect to the potential to combine the results of multiple sites or studies:
- When collecting data, identify individuals to the finest taxonomic level possible. Congeneric species, for example, may differ in indicator status (Weiss & Rice 2005; Nahmani et al. 2006) and may differ in nativity and other characteristics.
- Consider the proper exchangeable units for the permutations tests (Anderson & ter Braak 2003). Where appropriate, such as when a factor is nested within sites, sites can be analyzed separately and the results combined using meta-analytic techniques (Bakker 2008).
- Comparisons of ISAs from multiple studies require that the same typology be used in each study. Simple pairwise comparisons (e.g., grazed vs. ungrazed) are more likely to be comparable between studies than comparisons among multiple levels. Groups included in the typology should be described in detail.
- Ensure that the sample size within each group is sufficient to sample the vegetation. Doing so will also provide enough samples to permit an adequate number of permutations for assessing statistical significance.
- At a minimum, report the sample size and frequency of all species in all groups, so that the simplified IV can be calculated. Digital appendices and other repositories permit the archiving of these data in an accessible manner (Parr & Cummings 2005).
- To prevent publication bias in future meta-analyses (Gurevitch & Hedges 2001), publish data for all species, regardless of whether or not they were significant indicators.
What about TWINSPAN?
Prior to the development of ISA, the main statistical technique for identifying species associated with groups was Two-Way INdicator SPecies ANalysis (TWINSPAN) (Hill et al. 1975; McCune & Grace 2002, ch. 12). TWINSPAN is a divisive approach – it starts from the assumption that everything is in one group and then looks for ways to divide that group. However, several aspects of TWINSPAN greatly limit its utility:
- cannot be used for predefined groups
- performs poorly with > 1 gradient
- involves complex analytical details
- is based on Correspondence Analysis (CA) and therefore on chi-square distances
- requires “pseudospecies” to convert quantitative abundance data into qualitative presence/absence data (place abundances into classes and consider each class equivalent to a pseudospecies)
I have not seen TWINSPAN used in recent years.
Conclusions
ISA is an extremely powerful technique. It has high applicability for identifying species that differ between groups.
ISA differs from SIMPER in several ways. First, ISA provides more guidance about species that differ: if a species is an indicator of one group over the other we know that it is more abundant and frequent in the first group. In contrast, SIMPER quantifies how much a species contributes to the difference between groups but doesn’t indicate where the species occurs. Second, all sample units are considered simultaneously in ISA whereas the contribution of each species to each pairwise distance is calculated in SIMPER and then those contributions are averaged. This means that the connection to the actual dissimilarity between sample units is not as strong for ISA, though I have not seen explicit comparisons of ISA and SIMPER.
ISA was not designed to relate species to continuously distributed explanatory variables, though it has been adapted for that purpose as part of a technique known as TITAN.
Data adjustments remain important; these calculations will be affected by adjustments. You should use the same adjustments as in all other aspects of your analysis.
References
Anderson, M.J., and C.J.F. ter Braak. 2003. Permutation tests for multi-factorial analysis of variance. Journal of Statistical Computation and Simulation 73:85-113.
Baker, M.E., and R.S. King. 2010. A new method for detecting and interpreting biodiversity and ecological community thresholds. Methods in Ecology and Evolution 1:25-37.
Bakker, J.D. 2008. Increasing the utility of Indicator Species Analysis. Journal of Applied Ecology 45:1829-1835.
De Cáceres, M., and P. Legendre. 2009. Associations between species and groups of sites: indices and statistical inference. Ecology 90:3566-3574.
De Cáceres, M., P. Legendre, and M. Moretti. 2010. Improving indicator species analysis by combining groups of sites. Oikos 119:1674-1684.
De Cáceres, M., P. Legendre, S.K. Wiser, and L. Brotons. 2012. Using species combinations in indicator value analyses. Methods in Ecology and Evolution 3:973-982.
De Cáceres, M. 2023. Indicator Species Analysis using ‘indicspecies’. https://cran.r-project.org/web/packages/indicspecies/vignettes/IndicatorSpeciesAnalysis.html
Dufrêne, M., and P. Legendre. 1997. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecological Monographs 67:345-366.
Gurevitch, J., and L.V. Hedges. 2001. Meta-analysis: combining the results of independent experiments. Pages 347-369 in S.M. Scheiner and J. Gurevitch (editors). Design and analysis of ecological experiments. Oxford University Press, New York, NY.
Hill, M.O., R.G.H. Bunce, and M.W. Shaw. 1975. Indicator Species Analysis, a divisive polythetic method of classification, and its application to a survey of native pinewoods in Scotland. Journal of Ecology 63(2):597-613.
Martin, R.A., and S.T. Hamman. 2016. Ignition patterns influence fire severity and plant communities in Pacific Northwest, USA, prairies. Fire Ecology 12:88-102.
McCune, B., and J.B. Grace. 2002. Analysis of ecological communities. MjM Software Design, Gleneden Beach, OR.
Nahmani, J., P. Lavelle, and J.-P. Rossi. 2006. Does changing the taxonomic resolution alter the value of soil macroinvertebrates as bioindicators of metal pollution? Soil Biology and Biochemistry 38:385-396.
Parr, C.S., and M.P. Cummings. 2005. Data sharing in ecology and evolution. Trends in Ecology and Evolution 20:362-363.
Pillsbury, R.W., R.J. Stevenson, M. Munn, and I.R. Waite. 2019. Relationships between diatom metrics based on species nutrient traits and agricultural land use. Environmental Monitoring and Assessment 191:228. https://doi.org/10.1007/s10661-019-7357-8
Ricotta, C., S. Pavoine, B.E.L. Cerabolini, and V.D. Pillar. 2021. A new method for indicator species analysis in the framework of multivariate analysis of variance. Journal of Vegetation Science 32(2):e13013.
Severns, P.M., and E.M. Sykes. 2020. Indicator Species Analysis: a useful tool for plant disease studies. Phytopathology 110(12):1860-1862.
Smucker, N.J., E.M. Pilgrim, C.T. Nietch, J.A. Darling, and B.R. Johnson. 2020. DNA metabarcoding effectively quantifies diatom responses to nutrients in streams. Ecological Applications 30:e02205.
Weiss, J.M., and S.R. Reice. 2005. The aggregation of impacts: using species-specific effects to infer community-level disturbances. Ecological Applications 15:599-617.
Zald, H.S.J., B.K. Kerns, and M.A. Day. 2020. Limited effects of long-term repeated season and interval of prescribed burning on understory vegetation compositional trajectories and indicator species in ponderosa pine forests of northeastern Oregon, USA. Forests 11:834. https://doi.org/10.3390/f11080834
PS: Graphs of Indicator Value Contours
Here’s the code to create the above figure showing contours for Indicator Value based on different values for Aij and Bij.
temp <- expand.grid(Aij = seq(0, 1, 0.01),
Bij = seq(0, 1, 0.01))
temp <- temp %>%
mutate(IVij = Aij * Bij,
sqrt.IVij = sqrt(IVij))
IV.graph <- ggplot(data = temp,
aes(x = Aij, y = Bij, z = IVij)) +
geom_contour_filled() +
geom_contour(colour = "black") +
labs(title = "IndVal",
x = "Specificity (Aij)",
y = "Fidelity (Bij)") +
theme_bw() +
theme(text = element_text(size = 9))
sqrt.IV.graph <- ggplot(data = temp,
aes(x = Aij, y = Bij, z = sqrt.IVij)) +
geom_contour_filled() +
geom_contour(colour = "black") +
labs(title = "squareroot(IndVal)",
x = "Specificity (Aij)",
y = "Fidelity (Bij)") +
theme_bw() +
theme(text = element_text(size = 9))
ggarrange(IV.graph, sqrt.IV.graph, common.legend = TRUE)
ggsave("graphics/IndVal.contours.png",
width = 5, height = 3, units = "in", dpi = 600)
Media Attributions
- IndVal.contours
- Bakker.2008_Figure2

