2 Transcription, Translation
Session Level Objectives (SLOs): after completing the session, students will be able to:
SLO 5. Describe the basic ways in which microRNA (miRNA) molecules control gene expression.
Introduction
Common Themes of Information Transfer
DNA —replication—> DNA —transcription—> RNA —translation—> polypeptide
There are common themes in these reactions.
- Biological polymerization reactions always have an intrinsic directionality (polarity): In DNA replication and RNA transcription, the template strand is always read 3´-to– 5´, and the nascent chain is always synthesized 5´-to-3´. In protein synthesis (translation), the mRNA template is always read 5´-to-3´, and the nascent polypeptide is always synthesized N-to-C.
- The polymerase must be accurately positioned at a start site on the template. In each case this process is called initiation, and in each case initiation entails several regulated steps.
- The polymerase has an elongation cycle, which is what it sounds like. This is the major biosynthetic stage.
- A signal on the template signals termination of polymerization. Termination involves disassembly of the elongation machinery and the release of templates and products.
This is a general framework. We will not focus on every stage for each process, but rather on key stages that illustrate important concepts.
SLO 1. Explain why different genes are expressed at different rates, in different cells, at different times.
Most of the mammalian genome consists of non-coding DNA. A minority (~2%) of the human genome actually serves to encode mRNAs that are used as templates for protein synthesis. A similarly small fraction of the genome encodes other varieties of biologically important RNA molecules.
The fundamental question about gene expression is this: each of us has many different cell types, but only one genome. The different cell types are different because they make different proteins: muscle cells make contractile proteins; nerve cells have the enzymes needed to manufacture neurotransmitters; osteoblasts have the protein machinery needed to manufacture bone, and so on.
Moreover, even a single cell type needs to make different proteins at different times: we synthesize and secrete insulin (a peptide hormone) when we eat. We remodel entire tissues and organs throughout growth, in response to injury, and during pregnancy. How does this happen?
DNA —transcription—> RNA —translation—> polypeptide
The level of any given protein is a function of competing processes: synthesis and destruction. Protein synthesis requires an mRNA template, and the abundance of the mRNA encoding any given protein is also regulated by a balance of synthesis and destruction.
SLO 2. Describe the roles of mammalian RNA polymerases and key components in the RNA polymerase II (RNAP II) transcription preinitiation complex, including gene promoters and transcription factors.
RNA Transcription
The cell makes RNA molecules that do different things:
- mRNA is the template for protein synthesis (translation).
- tRNA and rRNA are core parts of the protein synthesis machinery.
- Diverse RNA molecules are involved in regulating gene expression and other processes. Examples include micro RNA (miRNA) and long non-coding (lncRNA). Many other examples are emerging.
The various RNAs are made by three RNA polymerase (RNAP) enzymes:
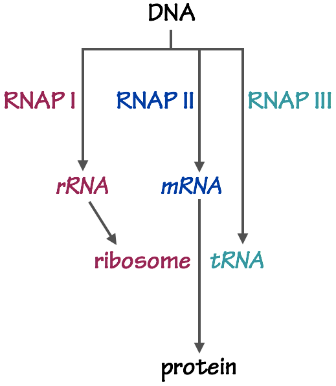
- RNAP I makes most of the ribosomal rRNA — the most important part of the ribosome, the enzyme that synthesizes polypeptides.
- RNAP II makes all of the messenger mRNA — the templates for polypeptide synthesis.
- RNAP III makes transfer tRNA — the carrier of activated amino acids for polypeptide synthesis.
We will focus on transcription by RNAPII, because its activity controls the levels of mRNA templates for protein synthesis. The underlying principles by which RNAP I and III operate are similar.
SLO 3. Describe a generic structure of a mammalian gene and its mRNA transcripts, including basic gene regulatory elements and processing reactions that precede export of an mRNA from nucleus to cytoplasm.
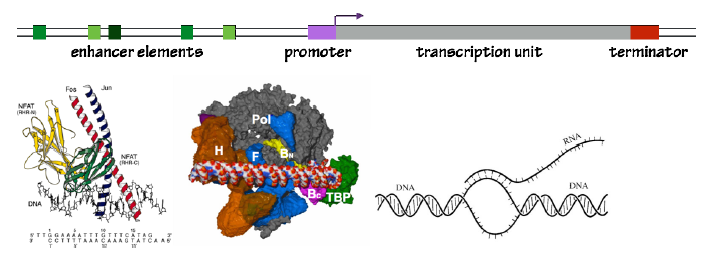
A gene contains two kinds of sequences:
- The transcription unit is the DNA sequence used as a template to synthesize RNA.
- Regulatory sequences tell RNAP where to initiate and terminate transcription. They allow cells to control which genes are actively transcribed (“expressed”), and which are silent. These sequences can be further sub-divided:
-
- The promoter sequence directs RNAP II and associated general transcription factors to the transcriptional start site.
- Enhancer sequences, usually 10-30 bp in length, bind transcription factors, or activator proteins, that instruct RNA polymerase to become active at the promoter. Each gene is controlled by different enhancer elements.
- Different cells contain specific sets of transcription factors. This is the main basis for cell-type-specific gene regulation! Enhancers can sit right next to the promoter, or tens of thousands of base pairs distant. Enhancers are usually upstream of the promoter but they can also be embedded within the transcription unit or even downstream of it.
- The terminator tells RNAP that it has reached the end of the transcription unit.
- There are other regulatory elements in the genome as well. For example, silencer sequences bind factors that, as the name suggests, decrease transcription. Insulator elements ensure that different genes are subject to independent regulation.
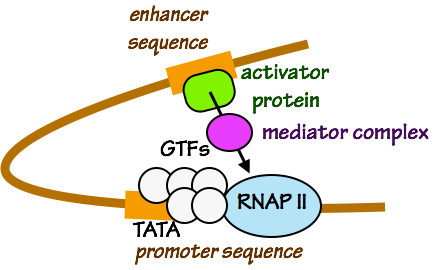
Fig. 3. Activation of RNAP by an activating transcription factor and a coactivator (in this example, “mediator complex”). The sequence TATA is often found in promoter regions. Enhancer sequences can be hundreds or thousands of base pairs distant from a promoter sequence.
Sequence of Events in Transcription:
- Genomic DNA is packaged into chromatin. Transcription is regulated in part by how densely packaged a given gene is, and hence, how accessible its regulatory sequence elements are. Later, we will discuss how DNA packaging is controlled.
- To initiate transcription of a gene, RNAP II must be directed to the promoter. This is done by the General Transcription Factors (Figs. 5 and 6). The GTFs recognize and bind to promoter sequences. They place RNAP II at the start site. GTFs then locally melt the DNA at the promoter, separating the two DNA strands to form a transcription bubble.
- The GTFs are “general” transcription factors because they are always needed for initiation of transcription. However, the GTFs do not have the ability to regulate when RNAP actually initiates RNA synthesis.
- In other words, GTFs are necessary but not sufficient for initiation of transcription.
- Transcription factors that bind to regulatory promoter, so intervening DNA must loop out in order for transcription factors, coactivators, and the initiation complex (GTFs + RNAP II) to touch one another. enhancer sequences are needed to activate transcription by RNAP II and the GTFs.
- Activating transcription factors “talk” to RNAP II and the GTFs by binding to coactivators that touch RNAP II and the GTFs (Fig. 3). Together, these events cause the pre-initiation complex — RNAP II and the GTFs — to initiate RNA polymerization. As we will see in a later session, transcription factors can also control chromatin structure.
- The chemistry of RNA polymerization is similar to the chemistry of DNA polymerization. The most important differences are:
- No primer is needed for RNA synthesis.
- NTPs are used for RNA synthesis, not 2-deoxy dNTPs.
- As RNAP II elongates the nascent mRNA chain, it moves along the template strand of the transcription unit (Fig. 3). The replication bubble moves as RNAP II “crawls” along the DNA template strand. In other words, the DNA double helix melts in front of RNAP II and re- hybridizes (anneals) behind it.
- When RNAP II reaches a terminator sequence (Fig. 3), the newly-synthesized RNA chain is released, RNAP is removed from the template strand, and the transcription bubble collapses.
The reason we care so much about the mechanics of RNAP II transcription is that this process controls which mRNA transcripts are produced, and in what abundance. This in turn controls the specific repertoire of proteins that can be made by each cell.
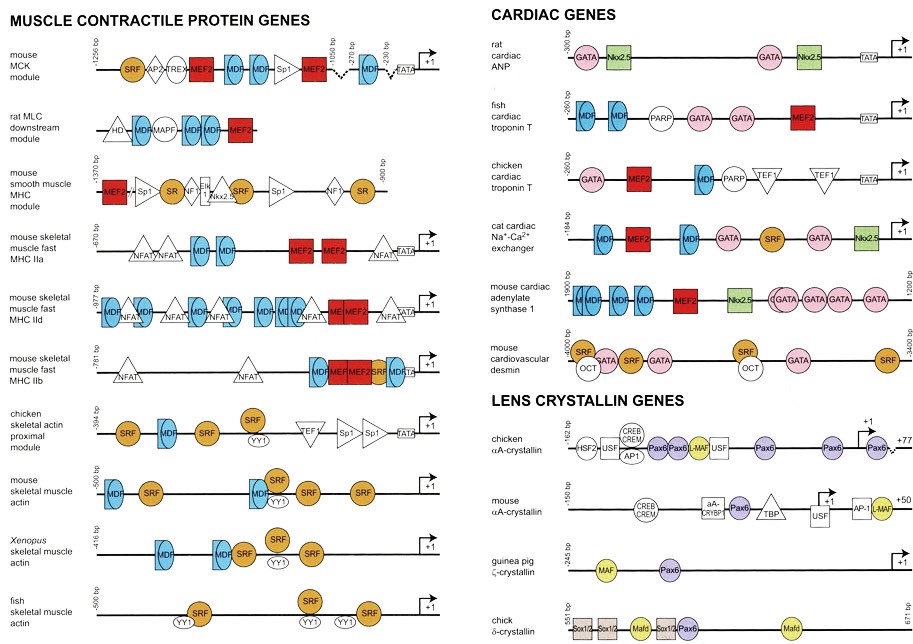
Fig. 4 shows the regulatory sequences of genes that encode some key proteins made only in specific kinds of cells: skeletal muscle cells, heart muscle cells, and cells in the lens of the eye.
Each type of DNA enhancer element sequence is recognized and bound by specific activating transcription factors — shown here by colored shapes. Humans have about 2,000 different transcription factors.
By producing specific combinations of transcription factors, each cell specifies which subsets of genes are actively transcribed, and in what quantities.
This concept is so important that it bears repeating:
The specific array of transcription factors, present in a given cell, shapes that cell’s pattern of gene expression — and thus, that cell’s overall protein complement, the cell’s identity (muscle, fibroblast, neuron, etc.) and its functional characteristics.
Another important point is that transcription factors are proteins. Consequently, the genes that encode transcription factors are themselves subject to transcriptional regulation. By transcribing and translating specific transcription factors the cell can execute temporary or stable programs of gene expression in response to developmental cues and other signals, such as food or infection.
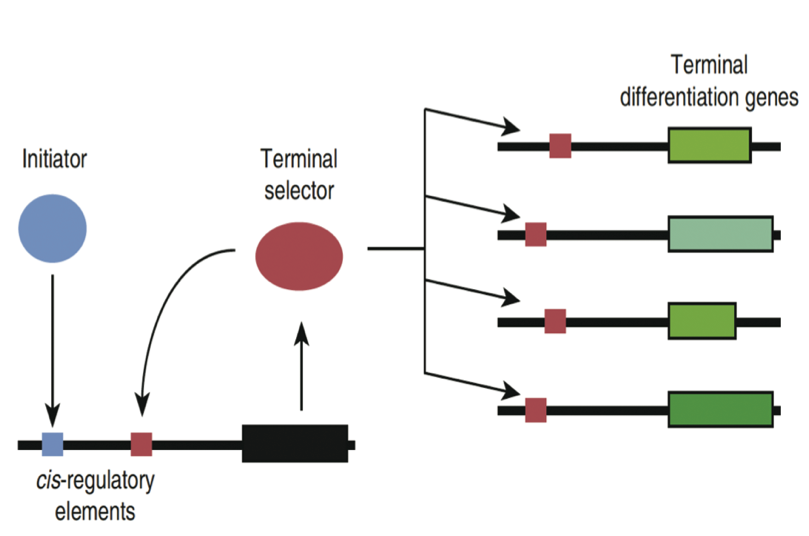
Fig. 5. Positive feedback loops maintain gene expression programs. Here, an “initiator” transcription factor binds an enhancer on the gene encoding a “terminal selector” transcription factor. When this gene is transcribed and the resulting mRNA is translated, the resulting protein binds to another enhancer in its own gene, and ensures that the terminal selector gene continues to be transcribed. The terminal selector also stimulates transcription of other genes needed for specific functions. Source: PNAS 110:7101
Processing reactions that precede export of an mRNA from nucleus to cytoplasm.
mRNA Processing and Export
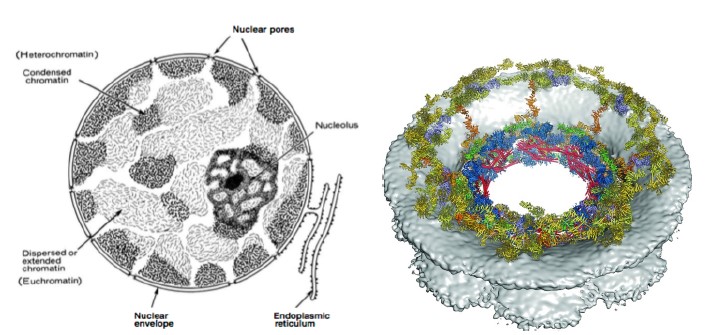
DNA replication and RNA transcription both occur in the cell’s nucleus. However, proteins are synthesized in the cytoplasm. To serve as templates for protein synthesis, mRNA molecules must be exported from the nucleus to the cytoplasm. This occurs at a special portal in the nuclear membrane, the nuclear pore (Fig. 6). The nuclear pore is an immense molecular assemblage that precisely controls the passage of mRNA and other macromolecules into, and out of, the nucleus. This is another theme that we will encounter again and again: biosynthetic products made in one cellular organelle are shuttled to another location — as with stations on an assembly line.
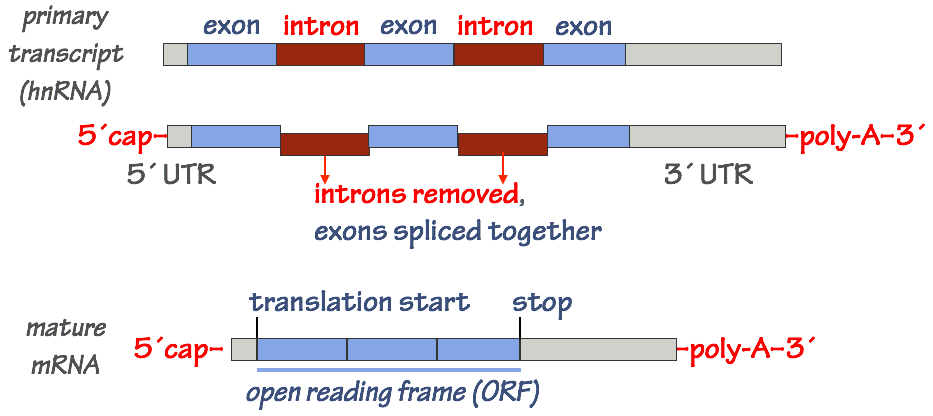
In the nucleus, the initial “raw” RNA transcript made by RNAP II must be processed. Only then is the mature mRNA exported from the nucleus to the cytoplasm, where it will be used as a template for protein synthesis.
- At the 5´end of the mRNA (the “beginning” of the transcript), a special nucleotide, 7– methylguanosine, is attached through a covalent bond. This is called the 5´ cap.
- Later, in the cytoplasm, the 5´ cap will tell the protein synthesis machinery that the RNA bearing the cap is a messenger mRNA — a template for protein synthesis — and not some other type of RNA.
- At the 3´ end of the transcript, a string of adenosine (A) nucleotides is added. This is the poly–A tail of the mRNA. The poly-A tail is constructed by a special enzyme (poly-A polymerase) through a non–templated polymerization process.
- The poly-A tail will signal export of the mRNA from nucleus to cytoplasm. It will also control the stability (the half-life) of the mRNA once it’s in the cytoplasm.
- The mRNA is spliced to remove introns and ligate (join) exons together. This step is somewhat involved and extremely important, so we’ll examine it in a bit more detail.
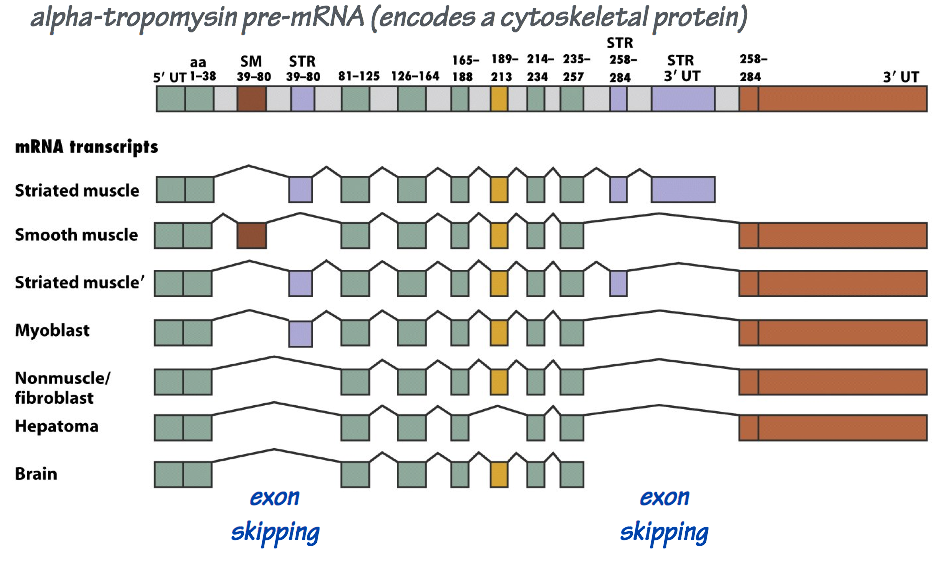
Differential mRNA splicing
The maturation of a large mRNA molecule may entail dozens of splicing reactions. In different cell types, these splicing reactions may be regulated so that not every exon ends up in each final, mature mRNA molecule. Consequently, a single transcription unit may encode more than one mRNA variant, with each derived from a different combination of exons (Fig. 8).
Differential splicing allows the ~20,000 protein-coding genes in the human genome to encode substantially more than 20,000 distinct mRNA templates and, thus, a much greater diversity of proteins.
To summarize:
- Transcription initiation controls how many mRNA transcripts get made.
- Different cells have different activating transcription factors.
- Different genes have different enhancers that bind different transcription factors.
- mRNA cap addition signals that the mRNA will be a template for protein synthesis.
- Differential splicing controls which exons are in the mature mRNA template, and thus the sequence of the resulting polypeptide.
- The poly-A tail (along with other features of the mRNA) controls nuclear export and the stability of the mRNA — how long it persists in the cytoplasm.
SLO 4. Outline the major steps of protein synthesis, including the roles of mRNA, tRNA, tRNA synthetases and ribosomes.
Here we summarize how polypeptide chains are synthesized and how they fold into their correct three-dimensional configurations.
The notes for this section begin with two charts: first, the assignments of RNA codons to amino acids (the genetic code); second, the chemical and physical properties of the 20 regular amino acids listed by features. This table also includes the rare amino acid selenocysteine, sometimes considered as the “21st amino acid,” although it is a modification of cysteine.
You do NOT need to memorize Tables 1 and 2! You do need to be able to apply their content.

Table 1
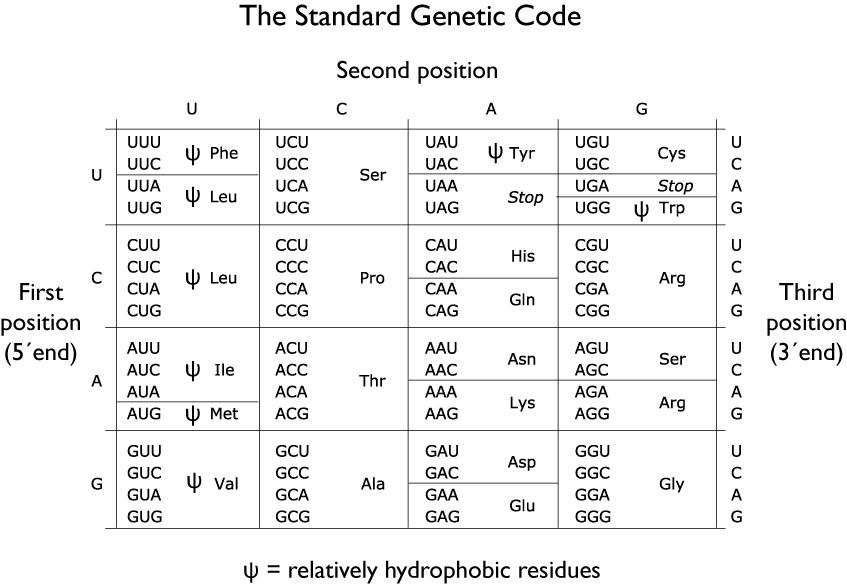
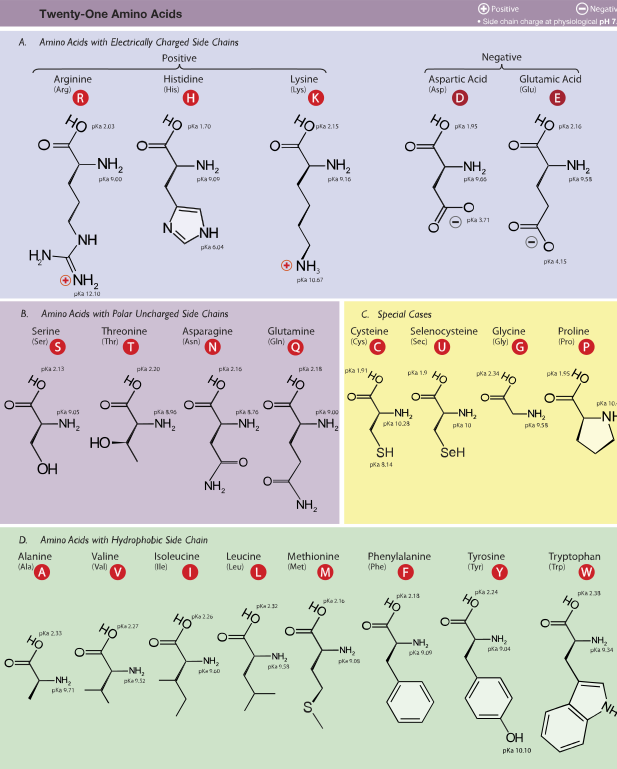 Table 2
Table 2
Source: Wikimedia
Protein Synthesis: the Genetic Code
Like nucleic acid synthesis, protein synthesis is a template-directed process. However, in protein synthesis, the process is a bit more complicated because the template does not directly interact with the polypeptide product. How this works has never been explained more plainly than by Francis Crick, in an astonishing proposal that he made in 1955:
…Each amino acid would combine chemically, at a special enzyme, with a small molecule which, having a specific hydrogen-bonding surface, could combine specifically with the nucleic acid template. This combination would also supply the energy necessary for polymerization… there would be 20 different kinds of adaptor molecule, one for each amino acid, and 20 different enzymes to join the amino acid to their adaptors…
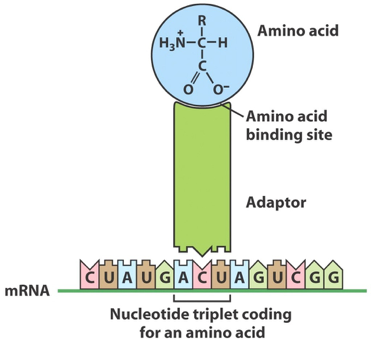
We now know that the “adapter” is tRNA. Crick’s proposal was correct in every detail save one: there are 61 possible combinations of 3-base mRNA codons (see Table 1), so there are more than 20 tRNA “adapters.” This system provides the biochemical basis of the genetic code — the rules through which each 3-base codon in an mRNA template specifies one specific amino acid.
tRNA and aminoacyl tRNA synthetase enzymes
-
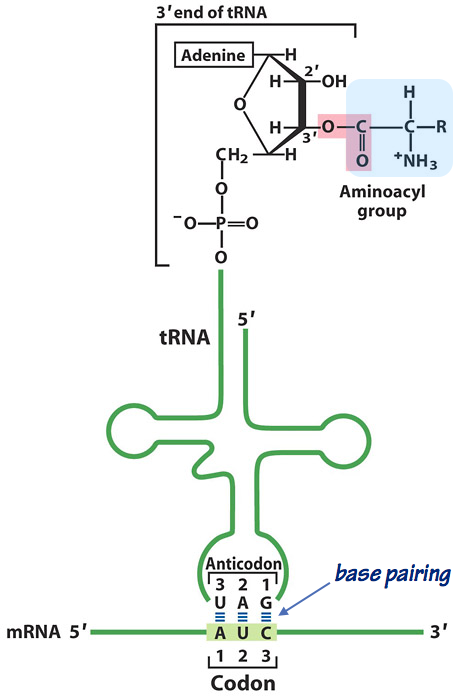
Fig. 10. An aminoacylated tRNA “reading” the genetic code. Each tRNA has 2 “business ends”:
- The anticodon pairs with the 3-base codon on the mRNA template. Look closely at the diagram (Fig. 13). Note that as in other nucleic acid hybrids, the two strands are antiparallel.
- The aminoacyl acceptor site is a terminal adenosine (A) nucleotide, where the carboxyl group of the amino acid is esterified to the 3´-OH of the adenine. Note that this is a relatively unstable, high–energy bond. It will make polypeptide elongation thermodynamically downhill, and hence favorable — exactly as predicted by Crick.
- Any given tRNA can (in principle) be esterified to any amino acid. However, the fidelity of translation depends absolutely on the accurate matching of a tRNA bearing a specific anticodon to the corresponding amino acid.
- The job of coupling each amino acid to its corresponding tRNAs is done by 20 different enzymes: the aminoacyl tRNA synthetases (also called aaRS enzymes). This is an absolutely key point: the specificity of the genetic code is controlled by the tRNA synthetases.
- The tRNA synthetases produce aminoacylated tRNA molecules in a two-step reaction (Fig. 11).
- First, an ATP is coupled to the amino acid to form an amino adenylate. That is, the amino acid is coupled to AMP. This is a high-energy activated intermediate.
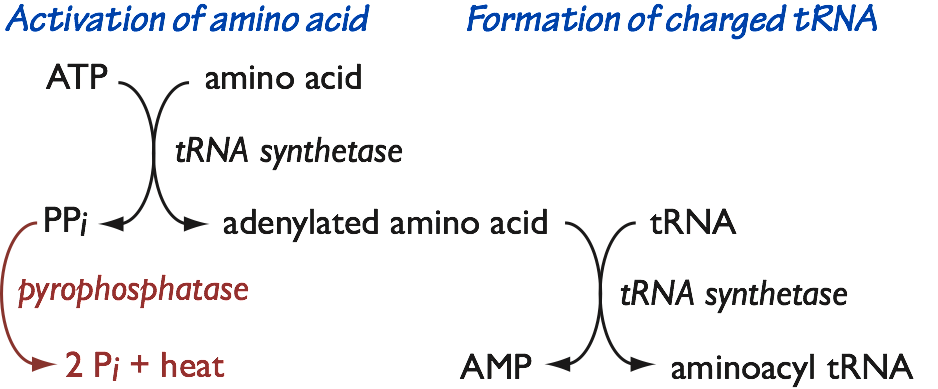
Fig. 11. Formation of aminoacyl tRNA. - In the first step, pyrophosphate (PPi) is also released. As we saw in DNA and RNA polymerization, pyrophosphate is immediately destroyed by pyrophosphatase, making the first sub-reaction an irreversible committed step.
- Second, the aminoacyl group is transferred to the tRNA. The products are an aminoacyl tRNA, and AMP. Because we start with ATP, and end up with AMP and two inorganic phosphates, coupling of an amino acid to tRNA has an energetic cost of 2 ATP equivalents.
- First, an ATP is coupled to the amino acid to form an amino adenylate. That is, the amino acid is coupled to AMP. This is a high-energy activated intermediate.
The Ribosome
The “polypeptide polymerase” is the ribosome, an enormous ribonucleoprotein complex. The ribosome has two subunits. Each subunit contains both RNA and many different polypeptides.
- The small subunit (it is not small, just not as big as the large subunit!) is the “decoding center.” The small subunit’s job is to match each codon on the template to a corresponding aminoacyl-tRNA. This is no easy task. There are 61 codons that specify 20 amino acids (Table 1), so a large majority of the aminoacyl-tRNA molecules that enter the ribosome must be rejected.
- When a correct codon-anticodon interaction is detected by the small subunit, the large subunit catalyzes the peptidyltransfer reaction — the chemistry of polypeptide elongation.
Remarkably, although each ribosome subunit contains both peptides and rRNA, both the decoding center within the small subunit, and the peptidyltransfer center in the large subunit, are made of ribosomal rRNA. The enzymatic core of the ribosome is a “ribozyme”. This is probably a relic of the ancient origin of ribosomes at the dawn of life, in the so-called RNA world.
- Initiation: the polymerase — the ribosome — must be placed precisely over the start codon on the mRNA template.
- Elongation: this is where template-mediated polymerization of the polypeptide occurs.
- Termination: A stop codon is identified, triggering release of the polypeptide and removal of the ribosome from the mRNA template.
Initiation of protein synthesis
As with DNA replication and transcription, initiation of protein synthesis is pretty complicated. And since the frequency of initiation controls the rate of protein synthesis, this step is also highly regulated.
Regulation occurs at both a global level (the cell asks how much protein synthesis it can support overall, given the available energy and resources), and for specific mRNA transcripts, which are translated with different efficiencies.
Steps in translation initiation
- The ribosome small subunit (Fig. 12, small brown oval) assembles with initiation factors.
- Together, they bind to the 5´cap of the mRNA.
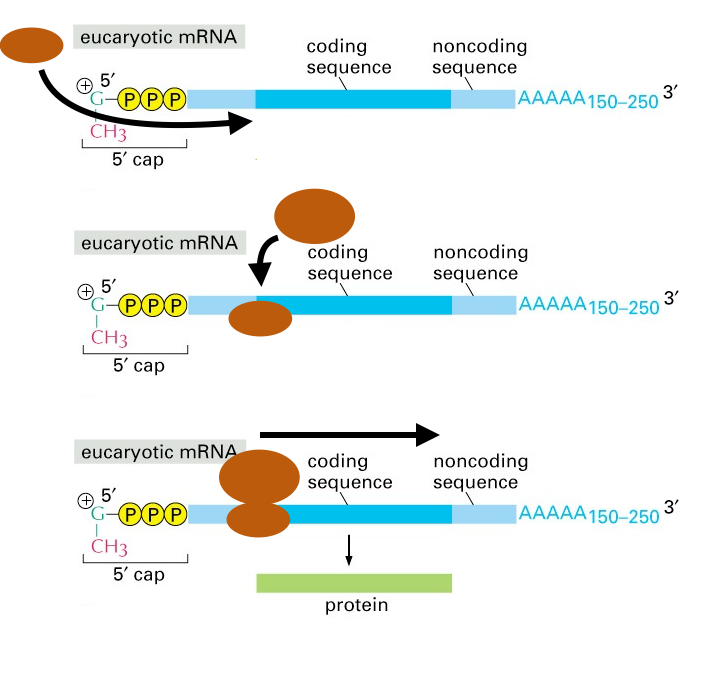
Fig. 12. Initiation of polypeptide synthesis by the ribosome. In this cartoon, the ribosome large and small subunits are the brown ovals. Translation initiation factors are not shown. Source: Alberts, Molecular Biol. of the Cell - The small subunit and initiation factors crawl along the mRNA from 5´-to3´, scanning the mRNA for a start codon.
- In most cases the start codon is AUG. If you look at Table 1, you’ll see that AUG encodes the amino acid methionine (Met, M). Thus, the first amino acid in a polypeptide is usually Met.
- Together, they bind to the 5´cap of the mRNA.
- Once the decoding center is accurately placed over the AUG start codon, the large subunit (in Fig. 12, the larger oval) docks onto the small subunit and mRNA, and the initiation factors fall off (dissociate). Now the elongation cycle can begin.
Two additional points about translation initiation must be emphasized.
First, as mentioned above, this step is highly regulated. Initiation factors are largely responsible for this regulation.
Second, the accuracy with which the ribosome’s small subunit is placed over the start codon is critical: a positional error of ± 1 or 2 nucleotides will put the mRNA transcript out–of–frame, and result in a totally different, and incorrect, polypeptide sequence. Similarly, if a mutation in the genome inserts or deletes one or two DNA bases in the coding region of a gene, the resulting mRNAs will contain frameshift errors. When this happens, every codon following the frameshift will be incorrectly decoded during translation. Most often this also results in early termination of synthesis.
Note: There are two major, important differences in mRNA structure and translation initiation between eukaryotes (humans included) and bacteria.
- In bacteria, mRNA molecules do not have a 5´ cap or 3´poly-A tail (Fig. 13). In addition, bacterial mRNA molecules often have a series of translation start sites, and a series of coding regions. Such an mRNA is referred to as “polycistronic.” In eukaryotes, a mature mRNA template usually has only a single st
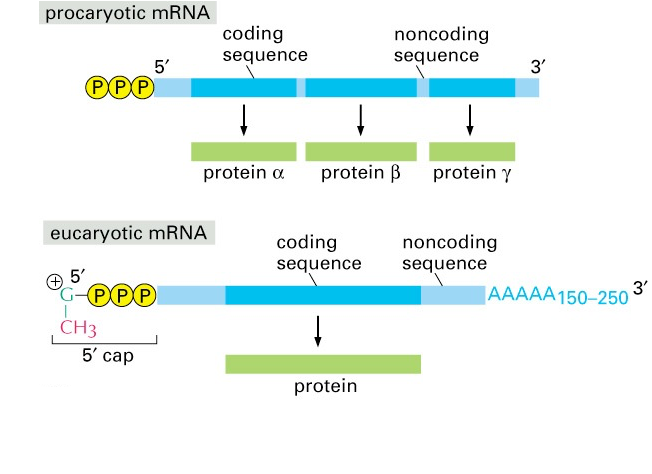
Fig. 13. mRNA structure in prokaryotes (bacteria) versus eukaryotes (including humans). Source: Alberts, Molecular Biology of the Cell art site and encodes only a single polypeptide.
- In bacteria, the first amino acid is usually a Met deriviative, fMet (formylmethionine). Peptides beginning with fMet are recognized by our innate immune system as a danger signal, because they can indicate an active bacterial infection. You will learn more about bacteria, danger signals, and innate immunity in the Infections and Immunity Block.
Polypeptide elongation
The assembled ribosome (large and small subunits) has three sites that can accommodate tRNA molecules: the A, P, and E sites. These names are shorthand for aminoacyl-tRNA, peptidyl-tRNA, and exit sites. It will become clearer in a moment why these names are used. The three sites are used in sequence. Here’s how it works (Fig. 14).
- At the A site, the ribosome samples incoming aminoacyl-tRNA (aa-tRNA) molecules. The ribosome is looking for an aa–tRNA with an anticodon correctly pairs with the mRNA codon positioned under the A site. Dozens of incorrect aa-tRNAs are rejected for each correct match.
- When a correct codon-anticodon match is identified, the ribosome initiates the peptidyltransfer reaction — the chemistry.
- The growing polypeptide chain, still esterified at its C (carboxyl)-terminus to a tRNA, resides in the P site.
- In the peptidyltransfer reaction, the nasent polypeptide is transferred from the peptidyl-tRNA sitting in the P site, to the aminoacyl residue sitting in the A site. This is counterintuitive. Inspect Fig. 14 to see how it works.
- Now the ribosome undergoes a ratchet-like twisting motion. This motion causes translocation of the mRNA, the peptidyl-tRNA, and the deacylated (discharged) tRNA. Now the peptidyl-tRNA is one residue longer, and in the P site. The deacylated tRNA is in the E site, where it is ejected from the ribosome. And the A site is unoccupied, ready to begin the cycle again for the next codon on the mRNA template.
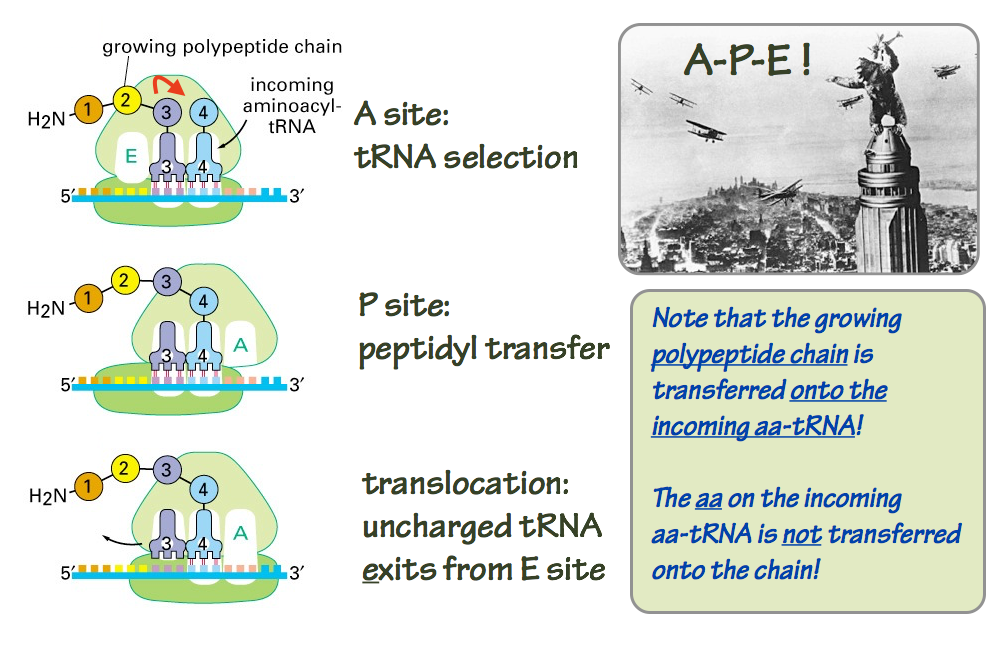
Fig. 14. Polypeptide elongation. Source: Alberts, Molecular Biology of the Cell
A final point about energy. Two ATP equivalents used to charge each aa-tRNA. This powers the peptidyltransfer reaction. However, additional ATP equivalents are consumed during the tRNA selection and translocation portions of the elongation cycle. In terms of both energy and materials, protein synthesis is very expensive. Many cell types use most of their energy on protein synthesis.
SLO5. Describe the basic ways in which microRNA (miRNA) molecules control gene expression.
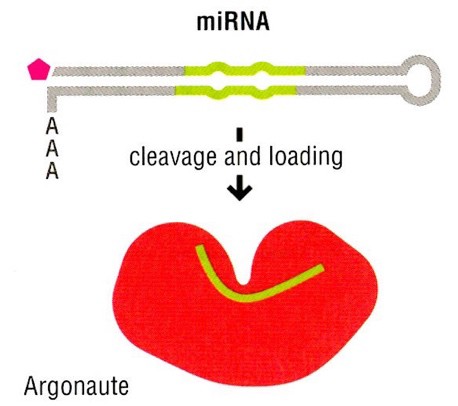
MicroRNAs (miRNAs) are post-transcriptional regulators of gene expression. More than 2,000 miRNAs have been annotated in human genome. 60% of all human genes are estimated to be regulated by one or more miRNA. miRNAs are short noncoding RNAs, usually about 22 nucleotides long. miRNA molecules are formed through two major pathways:
- Many miRNA precursors are coded as stand-alone genes, which can be transcribed by RNA polymerase II. Note that in the figure above, the miRNA is derived from a Pol II transcript with a 5´cap and 3´poly-A tail.
- As they are not coding sequences, miRNA precursors can also be derived from intron sequences that are embedded within other mRNA precursor transcripts. In these cases, splicing excises the intron and its miRNA from the coding exons of the mature mRNA.
miRNA does not function alone. miRNAs bind to the Argonaute family of proteins in the cytoplasm. miRNA-Argonaute complexes bind to specific mRNA transcripts via complementary hybridization between the miRNA seed region, and target sequences in the 3’UTR of the targeted mRNA transcript. In most cases, miRNAs function to repress (decrease) the production of specific sets of proteins. miRNA-Argonaute-mRNA complex can repress protein expression in different ways:
- destabilization of the mRNA via shortening poly (A) tail;
- inhibition of translation initiation;
- cleavage and degradation of the target mRNA.
Note that these mechanisms are post-transcriptional, meaning that they operate on mature mRNA molecules in the cytoplasm. In contrast, transcription factors act in the nucleus to control the rate at which different mRNA molecules are synthesized (transcribed) by RNA polymerase II. The first miRNA was discovered in the nematode C. elegans. Studies in humans have revealed that mutations in miRNAs can cause or contribute to various human diseases. For instance, mutation in miRNA-96 is linked to hereditary progressive hearing loss, and deletion of the miR-17~92 cluster causes skeletal abnormality and growth defects. Dysregulation of miRNA function is also implicated as a causative factor in several cancers.
SLO 6. Predict the molecular consequences of missense, nonsense, frameshift, synonymous and silent point mutations, and mutations caused by insertions, deletions, inversions, DNA expansions and amplification, in various parts of a gene or gene regulatory sequences.
It is important to be able to predict potential molecular consequences of various types of mutations as a foundation for understanding the physiological impacts of a genetic disorder.
Missense mutation: changes a codon resulting in a different amino acid in a protein encoded by that DNA sequence.
Nonsense mutation: introduces a premature stop (termination) codon in the DNA sequence encoding a protein, resulting in a truncated (shorter) polypeptide being synthesizes, as compared with the normal length.
Frameshift mutation: an insertion or deletion in the DNA sequence, that is not a multiple of three, changes the translational reading frame, resulting in an incorrect product of translation.
Synonymous mutation (silent or non-silent): a change in a codon does not result in a change in the amino acid encoded by the DNA sequence because of the redundancy in the genetic code. However this alteration could have other consequences to molecular processes that involve the presence of that sequence at that location in the DNA, such as transcription, splicing, mRNA export, translation and co-translational folding of the polypeptide. Consequently, a synonymous mutation may not be a “silent” mutation, where the mutation has no effect on production of the protein or resulting phenotype of the organism.
The consequence of different locations of mutations cannot be determined exactly, but predicting the effect of various locations of mutations aids understanding of the consequences of those mutations. For example, mutations in a promoter or enhancer can affect the rate of transcription while mutations within coding sequence of a protein-encoding gene can change the primary sequence of the protein product. Mutations at an exon-intron junction, as well as mutations in many coding and non-coding regions of a gene, can affect splicing of a primary RNA transcript.
Insertions and deletions (collectively called “indels”) or inversions could affect many molecular processes, depending on the DNA sequence involved and the breakpoints of the lesion. Gene amplification could cause over expression of a gene product, such as the HER2 gene in breast cancer.
When a repeated sequence appears as a greater number of iterations relative to normal, the primary sequence of the protein or the regulation of gene expression can be affected. Nucleotide repeat expansions such as those in Huntington disorder or Fragile X syndrome, will be discussed later in the course.
Feedback:
