8 Sickle Cell Disorder Mutations, Lab Techniques, Integration
fuerstpg
Sickle Cell Disorder Mutations, Lab Techniques, Integration
Note that the session learning objectives (SLOs) and information in this chapter was written by a former faculty member and does not parallel content presented in the current PowerPoint slide deck. A new chapter will be constructed in the future that is more directly relevant to the current content of this session.
Session Level Objectives (SLOs): after completing the session, students will be able to:
SLO 2: Explain how mutations in globin genes can reduce severe presentations of malaria.
Practice Quizzes:
SLO1. Describe the effect of common beta globin gene mutations on hemoglobin structure and function.
SLO5. Interpret results of electrophoretic assays used in diagnosing sickle cell trait and sickle cell disease.
Molecular Biology Techniques: Explain how different techniques are used to detect different types of biological molecules including northern, western and Southern blotting, ELISA, PCR, Sanger sequencing, RNAseq, (FACS) cell sorting and HPLC.
Synopsis: In this session we will consider mutations and molecular biology techniques and apply this knowledge to learn about sickle cell disease. Sickle cell disease is a debilitating disorder and sickle cell patients disproportionately suffer from health care disparities.
SLO1: Describe the effect of common beta globin gene mutations on hemoglobin structure and function.
There are two primary types of changes, or mutations, that occur in DNA. The first type of change is a base substitution. When a base substitution occurs, one letter of DNA is changed to another letter, for example A -> G. The second type of mutation is the insertion/deletion mutation, or indel mutation, in which the number of letters in a DNA sequence increases or decreases, for example GATC -> GAC (1 base deletion) or GATC -> GAATC (1 base insertion). A specific type of indel mutation is a base repeat expansion mutation, wherein a repeated number of bases is increased, for example CCG36 -> CCG42, which would be the addition of 6 new CCG repeats for a total of 18 new nucleotides.
Consequences of base substitution mutations
Base substitutions may occur within genes or in non-coding sequence outside of genes. Within genes, the base substitutions may occur within coding sequence or in non-coding sequence, for example within introns. The consequence of base substitutions varies widely depending on where the mutation is located.
Base substitutions in coding sequence can have four different effects on the coding sequence of the gene:
Synonymous mutation: The DNA changes, the mRNA changes, but the amino acid that is coded for does not change. An example would be GCA -> GCG, both codons of which code for alanine. If you consult the genetic code below you will notice that bases in the third position of the codon tolerate base substitutions.
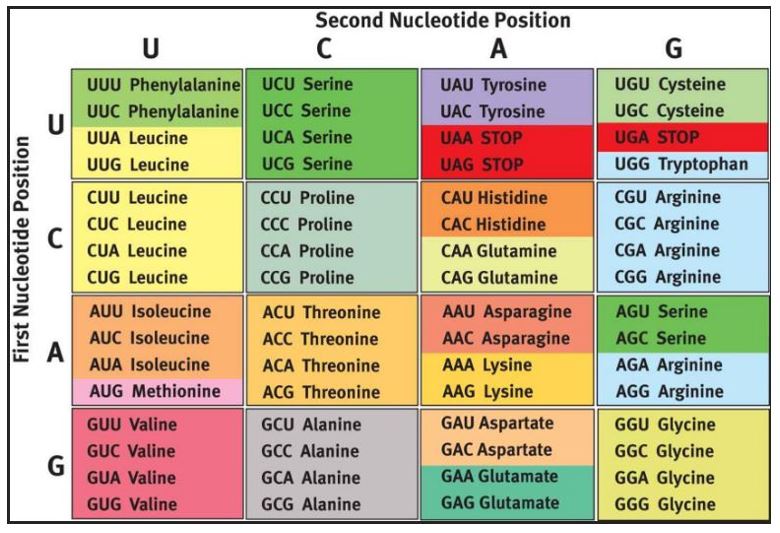
You will notice that some base substitutions in the third position will change the amino acid that is coded for and that some changes in other positions will not (see Arginine, Serine and Leucine, for example). In rare cases synonymous mutations can cause disease, for example if they disrupt normal splicing or regulatory sequences. Most synonymous mutations are silent mutations- that is there is no change in phenotype.
Nonsense mutation: A nonsense mutation changes an amino acid coding codon to one of the three stop codons, for example UGG -> UGA. This will terminate the protein early, which likely will have a profound effect on the resulting protein, depending on how close the new stop codon is to the native (normal) stop codon. Sometimes this will trigger a process called nonsense mediated decay, wherein mRNA with a premature stop codon is degraded. A premature stop codon introduced by an indel mutation is not a nonsense mutation! It is a premature stop codon introduced by an indel mutation. A premature stop codon introduced by an indel mutation can however result in the process of nonsense-mediated decay of the mRNA transcript.
Missense mutation: A missense mutation is a base substitution in which the amino acid that is coded for changes, for example AGA -> AGC. Some missense mutations have little or no effect on protein structure and function, for example a mutation that changes leucine to isoleucine may not have a detectable impact on protein structure and function. These are conservative missense mutations. A missense mutation that changes the class of amino acid or changes an amino acid with special properties is more likely to be a non-conservative deleterious mutation, for example the mutation GAA (glutamate) -> CAA (glutamine) will change the charge of the amino acid that is coded for. The mutation that results in sickle cell disease, GAG -> GTG (glutamate -> valine), is a non-conservative deleterious missense mutation.
Read through mutation: A rare type of base substitution changes a stop codon to an amino acid. An example would be TGA -> CGA (stop -> arginine). Read through mutations will result in longer proteins. Translation will continue until a stop codon is encountered in what would otherwise be the 3’ UTR.
Base substitutions in non-coding sequence: Changing a nucleotide base outside of coding sequences is likely to have no detectable outcome but if the base substitution is in regulatory sequence the change could be very deleterious. Examples include intron regulatory sequences, promotors and miRNAs. The vast majority of non-coding sequence does not play a regulatory or other role.
Consequences of insertion deletion (indel) mutations
Indel mutations in coding sequence: Indel mutations add or remove bases, hence the name. The impact of indel mutations in coding sequences depends on whether the mutation is in frame and whether it preserves the reading frame.
If the indel mutation is divisible by three and is in frame it will preserve existing or remaining codons and insert or delete codons.
In frame and preserves reading frame insertion example:
GAA CGC AUG TTT CCA ACC TCC TCC Codons (wild type)
E R M F P T S S Coded Amino Acids
GAA CGC AUG ACA GAC TTT CCA ACC TCC TCC Codons (mutant)
E R M T D F P T S S Coded Amino Acids
Out of frame but preserves reading frame insertion example:
GAA CGC AUA CAG ACG TTT CCA ACC TCC TCC Codons (mutant)
E R I Q T F P T S S Coded Amino Acids
It should also be noted that an indel mutation that preserves the reading frame can introduce a stop codon, for example if the introduced nucleotides code for a stop codon.
If the indel mutation is not divisible by three it will not preserve the reading frame of the transcript. In this case there is little difference if the mutation is in or out of frame- both examples will be very deleterious.
In frame does not preserves reading frame insertion example:
GAA CGC AUG ACA ACT TTC CAA CCT CCT CC Codons (Mutant)
E R M T T F Q P P Coded Amino Acids
Out of frame does not preserves reading frame insertion example:
GAA CGC AUA CAA CGT TTC CAA CCT CCT CC Codons (Mutant)
E R I Q R F Q P P Coded Amino Acids
It is likely that this type of mutation will introduce an early stop codon, but in some cases the indel mutation might cause translation to miss the native (normal) stop codon and code for a longer protein. This can occur if the mutation is close to the stop codon.
Indel mutations in non-coding sequence: Similar to base changes outside of coding sequences, indel mutations in non-coding sequence are much less likely to have a detectable outcome. If the indel is in regulatory sequence the change could be very deleterious however. Examples include intron/exon junctions and intron regulatory sequences, promotors and miRNAs. A specific example is the CGG repeat in Fragile X that results in methylation and inactivation of gene expression. Such nucleotide expansion repeat disorders affect the nervous system and will be covered in more detail in genetic and pathology sessions. The vast majority of non-coding sequence does not play a regulatory or other role however and the indel mutation in non-coding sequence is more likely to not have an impact because of this.
Gene duplication and deletion indel mutations: Sometimes indel mutations are very large and result in the duplication or deletion of an entire gene or genes.
Silent mutations: Any type of mutation can be a silent mutation (also referred to as a neutral mutation) – a mutation which does not result in a change to phenotype.
SLO 2: Explain how mutations in globin genes can reduce severe presentations of malaria.
See PowerPoint slide deck for contents of this SLO. Discussion in this chapter will be developed in the future.
SLO 3. Explain how mutations in globin genes can result in anemia and pain associated with sickle cell disease (SCD).
See PowerPoint slide deck for contents of this SLO. Discussion in this chapter will be developed in the future.
SLO 4. Describe various treatments for sickle cell disorder including gene addition and gene editing technologies.
See PowerPoint slide deck for contents of this SLO. Discussion in this chapter will be developed in the future.
SLO 5. Interpret results of electrophoretic assays used in diagnosing sickle cell trait and sickle cell disease.
Sickle cell disease is a type of hemoglobinopathy that is caused by a mutation in b-hemoglobin. b-hemoglobin combines with a-hemoglobin in a 2:2 ratio to form adult (A) hemoglobin. Note the Greek letters denote the individual protein chains while the Latin letters denote the hemoglobin molecules made up of four hemoglobin chains (Figure 3):
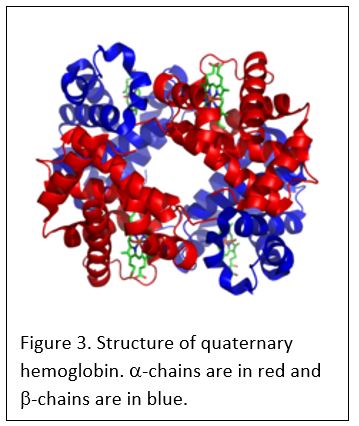
alpha-hemoglobin: one of two molecules in hemoglobin. Combines with b or g hemoglobin to form hemoglobin tetramers.
beta-hemoglobin: the adult hemoglobin molecule that combines with a hemoglobin.
gamma-hemoglobin: the fetal hemoglobin molecule that combines with a hemoglobin.
A-hemoglobin: Adult hemoglobin composed of two a and two b hemoglobin subunits (a2b2).
F-hemoglobin: Fetal hemoglobin composed of two a and two g hemoglobin subunits (a2g2).
There is a developmental switch from A to F type hemoglobin early in postnatal life. In fact some therapies for sickle cell disease try to reactive production of fetal F type hemoglobin.
The primary disease causing mutation responsible for sickle cell disease is a missense mutation that results in a substitution of valine in place of glutamic acid at amino acid #6. Valine is a hydrophobic amino acid and the mutation causes hemoglobin molecules to form chains because of hydrophobic interactions between valine and other hydrophobic amino acids in other b-hemoglobin molecules. A similar mutation, glutamic acid at position 6 -> lysine is responsible for hemoglobin C disease. Note, in this nomenclature the first methionine of the protein, which is removed during translation of many but not all proteins, is not counted. Hemoglobin C disease is mild- the main complication of the mutant allele is that it can combine with the hemoglobin S mutation to cause sickle cell disease.
S-hemoglobin: Adult hemoglobin composed of two a and two b hemoglobin subunits (a2bE>V2).
C-hemoglobin: Adult hemoglobin composed of two a and two b hemoglobin subunits (a2bE>K2).
Note: hybrid hemoglobin tetramers can form that are (a2bbE>V). These hybrid hemoglobin molecules actually inhibit sickling by blocking the chain formation that leads to sickling of red blood cells.
Sickle cell and hemoglobin C diseases are recessive:
Alleles Outcome
bE6->V/ bE6->V Sickle cell disease
Hemoglobin S only
bE6->K/ bE6->K Hemoglobin C-disease
Hemoglobin C only
bE6->V/ bE6->K Sickle cell disease
Hemoglobin C, Hemoglobin S
b/bE6->K Hemoglobin C trait
Hemoglobin A, Hemoglobin C
b/bE6->V Sickle cell trait
Hemoglobin A, Hemoglobin S
b/b Typical
Hemoglobin A only
ADDITIONAL DISCUSSION OF ANALYTICAL TECHNIQUES IN MOLECULAR BIOLOGY
Many of the techniques we will briefly review utilize electrophoresis (Figure 2), the process of separating molecules based on size and or charge through a matrix (agarose, acrylamide etc.) by applying an electrical charge across the matrix. The molecules that migrate can be stained with a variety of dyes and other labels to visualize them.

Other techniques make use of antibodies that are specific for (typically) a protein antigen. Antibodies may be polyclonal (for multiple B-cells) or monoclonal (from a single B-cell lineage). Antibodies are made by injecting the antigen into an animal that will have an immune response to the antigen and then isolating the antibodies or B-cells from the animal.
What follows is a very brief synopsis of a variety of molecular biology techniques that you will encounter in the curriculum.
Southern Blot: Like Northern blot but for DNA. DNA molecules are typically cut with a restriction enzyme before running them on a gel matrix by electrophoresis. Southern was a person, which is why Southern is capitalized.
Northern Blot: RNA molecules are separated by electrophoresis. Typically a radioactive complementary DNA or RNA molecules are used to detect and identify the presence, size and abundance of a specific type of RNA molecule. This technique is not frequently used anymore because of the advent of RNAseq.
Western Blot: Protein samples are separated by electrophoresis and specific protein molecules are detected with an antibody.
PCR (Polymerase Chain Reaction): Short DNA oligo primers are used to selectively amplify a DNA sequence with repeated cycles of strand denaturation and DNA synthesis*.
PCR: Short DNA oligo primers are used to selectively amplify a DNA sequence in polymerase chain reaction. The DNA may then be run or a gel and visualized, sequenced or used for a downstream application.
RT-PCR (reverse transcription-PCR): PCR but using cDNA as a template. cDNA is generated by reverse transcription of RNA. The COVID-19 PCR test is an RT-PCR test. Reminder- cDNA is generated by reverse transcribing RNA, typically mRNA. This differs from genomic DNA (gDNA) which would include intron sequences that are spliced out of mRNA molecules.
ELISA: Enzyme-linked immunosorbent assay. The ELISA test is a useful and common test to detect the presence of a specific antigen or antibodies to a specific antigen. ELISA tests are rapid tests and very frequently used clinically or over the counter for home use. Example: over the counter pregnancy tests are a type of ELISA test. ELISA tests involve incubating a small plastic well with solutions containing a mix of antigens and or antibodies. If the antibodies and antigens interact then they will remain in the well during washing steps. Otherwise, they will be flushed out of the well when the wells are washed.
There are several types of ELISA test:
Direct ELISA: An antigen is immobilized on a well. Antibody samples are labeled with an enzyme or fluorescent tag. The test measures if the antibody binds to the antigen. This can be used to measure if there was an immune response to an antigen.
Indirect ELISA: Like the direct ELISA an antigen is immobilized on a well. The well is incubated with an antibody (the “primary” antibody). A second “secondary” antibody that is labeled with an enzyme and binds to the primary antibody is then used to determine if the primary antibody bound to the antigen. The primary antibody binds to antigen the secondary antibody will bind to the first antibody and the result will be a positive reaction. If the primary antibody does not bind to the antigen then the secondary antibody will be washed out, resulting in a negative result.
Sandwich ELISA: A well is coated with an antibody specific for an antigen. If the antigen is present it will bind to this antibody and remain attached to the well during washes. A second labeled antibody that also binds the antigen is then added. If there is antigen “sandwiched” between the two antibodies a positive result will occur. If there is no antigen the labeled second antibody will not stick in the well after washes.
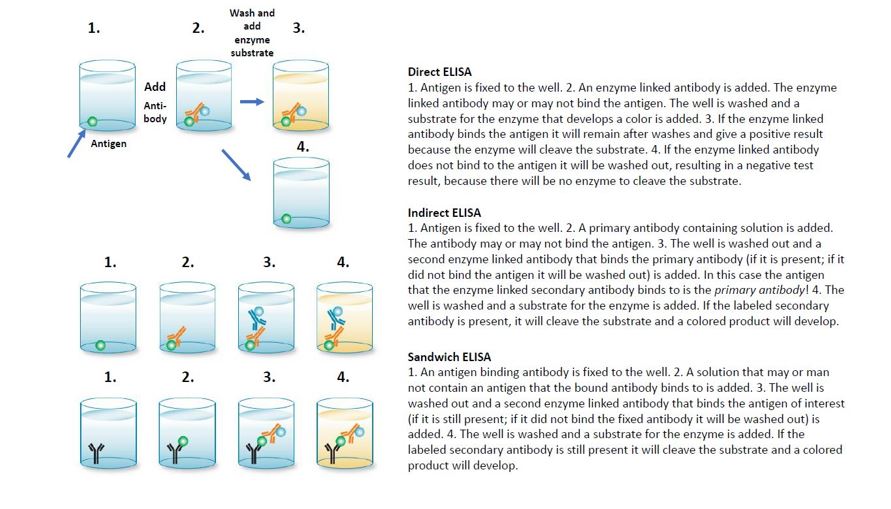
Competitive ELISA (not shown): ELISA tests can be modified to include a known amount of a competitor antibody or antigen. This can permit one to measure how much antigen or antibody there is in a solution.
Sanger Sequencing: Sanger sequencing, named after a scientist, is a very accurate “gold standard” method of sequencing a single purified population of DNA, which are typically generated by PCR.
Next generation sequencing: A lower accuracy (compared to Sanger sequencing) but high throughput method of sequencing an entire population of nucleic acid molecules. This technique has largely supplanted techniques like microarrays and northern blots.
FACS/flow cytometry: Fluorescent Activated Cell Sorting. A cell population is labeled, for example with a fluorescent antibody to CD4, and counted and or sorted on a cell sorter. This technique can be used to detect specific populations of immune cells based on their expression of specific receptors.
HPLC: High performance liquid chromatography: samples are separated based on their hydrophobicity and similar properties. The nature of some molecules can be detected based on their very specific sizes, or the sizes of their breakdown products, after separation by mass spectrometry.
Histochemistry/Cytochemistry: A technique by which tissues (histo) or cells (cyto) are probed and marked with specific labels, most often antibodies. When antibodies are used this is referred to as immunohistochemistry (IHC) or immunocytochemistry (ICC). The labels can be used to identify if a cell type expresses a specific protein, for example estrogen receptor/progesterone receptor testing of breast cancer biopsies.
Feedback:
