1 Nucleic Acids, Protein Building Blocks
Session Level Objectives (SLOs): after completing the session, students will be able to:
SLO 1. Explain the “Central Dogma” of molecular biology.
SLO 4. Explain how cells overcome three major challenges to replicate their genomes.
SLO 1. Explain the “Central Dogma” of molecular biology.
Sequence Information and the “Central Dogma” of Molecular Biology
The linear sequence of almost all polypeptides (there are a few exceptions) is stored, in encoded form, in the DNA of our genome. The flow of sequence information occurs whenever DNA, RNA, or protein polymers are synthesized:
DNA —replication—> DNA —transcription—> RNA —translation—> polypeptide
A critical point: each of these processes entails a series of chemical reactions. Each reaction is catalyzed by specific enzymes and is controlled by the laws of statistical thermodynamics. Consequently, biological information transfer processes are never error-free. They cannot be. Consequently, cells spend enormous energy, materials, and time to reduce and cope with errors in nucleic acid and protein synthesis. When these tactics fail, the consequences can be devastating.
A significant portion of this course will focus on errors in biological information transfer.
On the flip side, sequence changes (through DNA replication errors and other mutational processes) are responsible for all the richness and splendor of human genetic diversity. Understanding this diversity is essential to understanding and treating human disease and will only become more important as we are deluged with human DNA sequence data.
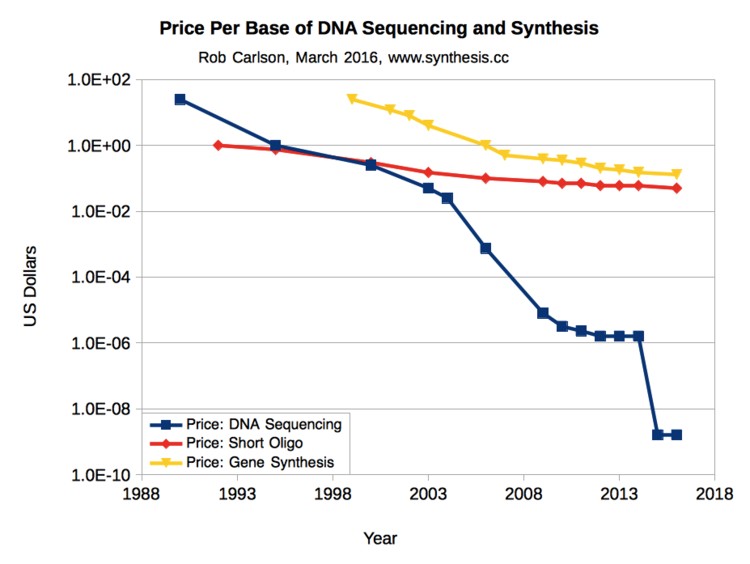
Fig. 1 shows that the cost of DNA sequencing has dropped faster than the price of electronic integrated circuits dropped from the 1970s to the present (Moore’s Law). In 2016 it cost about $1000 to sequence a human genome.
At present, you won’t see much DNA sequence data used in most clinical settings. But by the time you complete your medical training, the cost will have fallen to only a fraction of the current $1000 per genome — comparable to many standard lab tests.
Thus, it is essential that you should obtain a working understanding of human genetic variation and its consequences for health and disease.
SLO 2. Describe the general structure of nucleotides and consequences of nucleic acid base deamination.
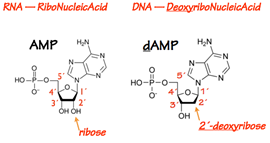
The protomer of a DNA or RNA polymer is the nucleotide (Fig. 2). A nucleotide contains a base, a 5- carbon (pentose) sugar, and one or more phosphate groups. If the sugar doesn’t have a phosphate group on it, the pentose-base unit is called a nucleoside.
In RNA the pentose is ribose. In DNA the pentose is 2´-deoxyribose — ribose lacking a hydroxyl at its 2´position.
To each sugar is attached a base (Fig. 3). The base is always attached at the 1´position of the pentose sugar through a glycosidic bond.
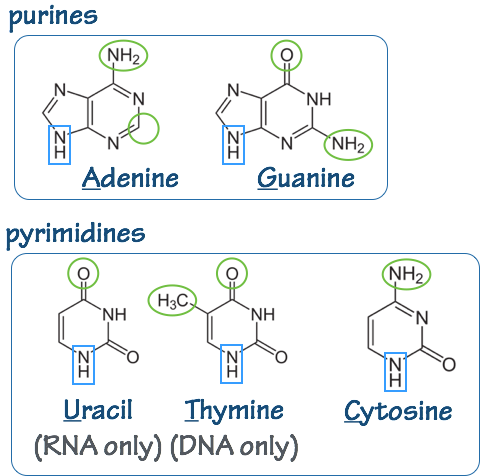
In DNA the bases are: adenine (A), thymine (T) guanine (G), and cytosine (C). In RNA, thymine (T) is replaced by uracil (U).
The nucleotides (base-sugar-phosphate) are called: adenosine (A), guanosine (G), thymidine (T), cytidine (C), and uridine (U). Often the names are written to indicate the phosphorylation state: Adenosine diphosphate (ADP), etc.
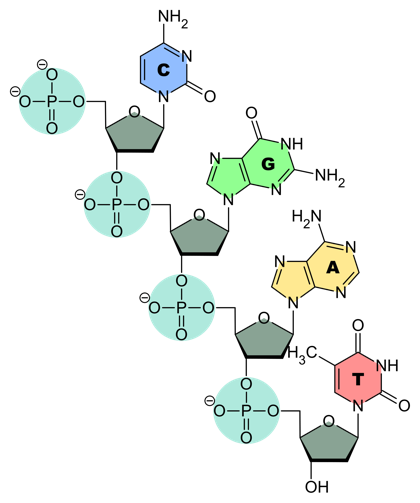
DNA and RNA chains are strings of linked nucleotides. Each chain (Fig. 4) consists of a backbone made out of alternating sugar and phosphate groups. Put a bit differently, the pentose sugars are linked by phosphodiester bonds.
As with proteins, DNA and RNA chains have polarity. This is defined by the orientation of the pentose sugar: one linking phosphate is attached at the 3´ position on the pentose, and one is attached at the 5´ position (see Fig. 3 and compare to the backbone in Fig. 4).
In biological polymerization reactions, nucleotides are always added at the 3´ end of an elongating chain. That is, the chain is polymerized 5´-to-3´. This leads to a convention: we write DNA and RNA sequences 5´-to-3´— unless explicitly specified otherwise.
SLO 3. Describe DNA and RNA secondary structure, polarity, and forces that stabilize the DNA double helix, including the role of water.
Strands of DNA or RNA can hybridize (anneal) to form double–stranded structures like the familiar DNA double helix.
- Specificity in hybridization is provided by base–pairing. A pairs with T (or U), G with C. Accuracy in pairing is promoted by favorable, non–covalent hydrogen bonds and by shape complementarity.
- The two pentose-phosphate backbones are at the exterior of the double helix. This makes sense: the sugars are very polar, and every phosphate group carries a negative charge. Both sugar and phosphate are hydrophilic — they like to interact with water.
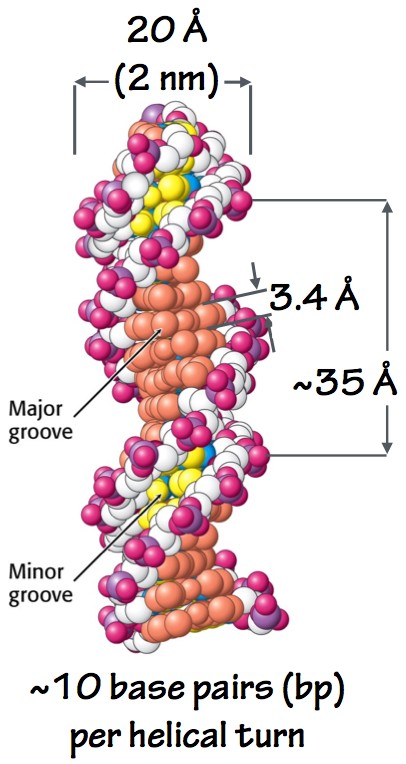
Fig. 5. DNA double helix. - The two sugar-phosphate backbones run antiparallel, like the traffic on a two-way street: one strand runs 5´-3´, and the other runs 3´-5´.
- Consecutive base-pairs stack like plates at the center of the helix, so close together that water is excluded. This also makes sense: the bases are flat and relatively hydrophobic. Their flat surfaces favorably interact (stack) with one another and are shielded from aqueous solvent. This also protects the bases from certain kinds of chemical attacks.
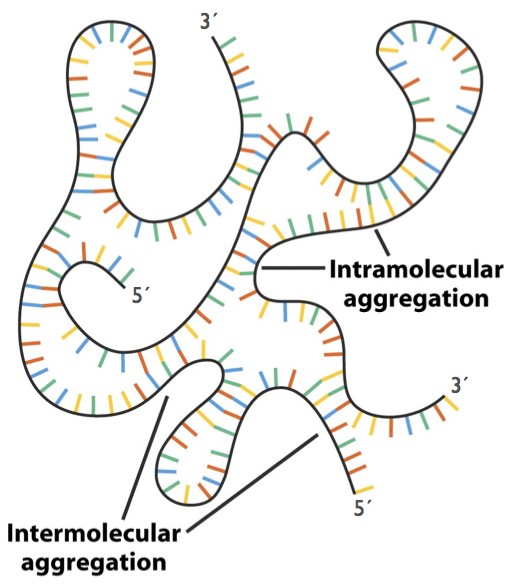
Fig. 6. Hybridization within and between RNA strands. Note that the backbones in hybridized regions are antiparallel. - To summarize: the stability of a double helix is controlled by base-pairing and also by other forces: separation of the negatively- charged phosphates, favorable stacking interactions between the flat bases, and the resulting shielding of the hydrophobic bases from the aqueous solvent.
- The double helix has a minor groove and a major groove (Fig. 5). The major groove is critical: it allows proteins to touch the bases and “read” the DNA sequence, as if by braille. Thus, regulatory proteins can identify and bind to specific short DNA sequences, without pulling the two strands apart.
- RNA can hybridize with RNA, or with DNA. In RNA biology, hybridization between complementary sequences on a single strand is of special importance (Fig. 6). Hybridization allows the formation of hairpin structures with short regions of double helix. These secondary structure elements can combine to generate complex tertiary structures including tRNAs and ribosomes, which are critical in protein synthesis.
- DNA and RNA diagnostics including microarrays and in situ hybridization are based on sequence-specific hybridization of short oligonucleotide probes to DNA or RNA analytes (samples).
SLO 4. Explain how cells overcome three major challenges to replicate their genomes
DNA replication and RNA transcription are chemical reactions that involve information transfer.
- To provide a template for DNA replication or RNA transcription, the DNA double helix must be locally pulled apart. Separation of the two strands is called melting or denaturation.
- A highly specialized DNA or RNA polymerase enzyme moves along a template strand. The polymerase elongates the nascent chain by testing the base-pairing of incoming nucleotides with the template.
- If the base-pairing is correct, the enzyme triggers the chemistry: the incoming nucleotide is added to the 3´end of the nascent chain. (Fig. 7).
- To power polymerization, the incoming nucleotides are NTPs (nucleotide triphosphates, for RNA) or dNTPs (deoxynucleotide triphosphates, for DNA):
…pNpN-3´+ NTP —polymerase—>…pNpNpN-3´+ PPi
Each lower-case “p” in the above scheme is one phosphodiester bond. The product of the reaction is a nascent chain with one additional nucleotide residue. A molecule of inorganic pyrophosphate (PPi = (P2O7)4–) is evolved.
- The polymerization reaction is potentially reversible. To make the reaction irreversible, the enzyme inorganic pyrophosphatase destroys the evolved pyrophosphate in a highly favorable, effectively irreversible, reaction:
PPi + H2O —pyrophosphatase—> 2 Pi + heat
In later sessions, we will see the destruction of pyrophosphate used to make additional metabolic reactions, such as protein and lipid synthesis, irreversible.
-
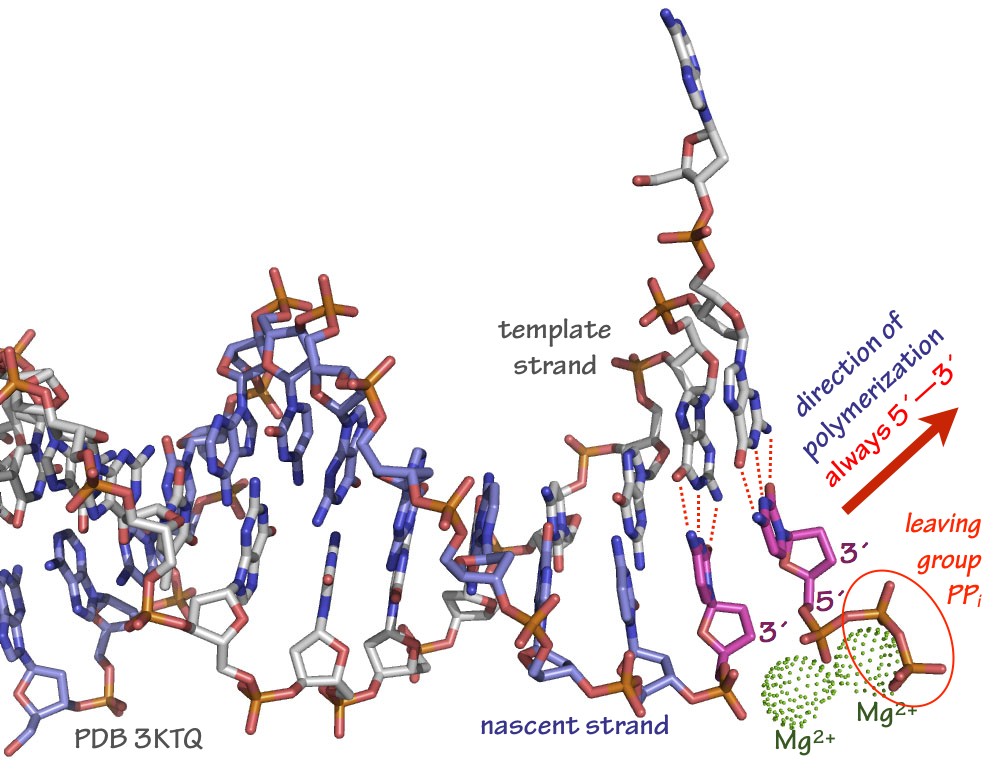
Fig. 7. Arrangement of template strand, incoming dNTP (in this case, dCTP), and stabilizing Mg2+ ions in a typical DNA polymerase active site. The polymerase itself is not shown in this rendering. Source: Merz, based on PDB 3KTQ In most cases the polymerase enzyme remains tightly bound to the template and nascent strands, and the elongation cycle begins again. The ability of an enzyme to catalyze many polymerization cycles without falling off (dissociating) from a template is called processivity. Some DNA and RNA polymerases have processivities of a million bases or more.
The main polymerases we’ll think about in this course are the enzymes that catalyze DNA replication and RNA transcription. However, other DNA and RNA polymerase enzymes exist. There are RNA polymerases that use RNA as a template, and DNA polymerases that use RNA as a template (“reverse transcriptases”). Our cells use these alternative polymerases for specialized housekeeping functions such as telomere maintenance. RNA viruses and retroviruses use these classes of enzymes in their infection and replication cycles, as you’ll see in the Infection and Immunity block.
To replicate its genome, the cell must overcome several challenges:
-
- There are two DNA strands to be replicated.
- The two strands run in opposite directions (they’re antiparallel).
- Replication must be accurate.
- Enormous amounts of DNA are replicated: 6 Gbp/cell (= 6×109 base pairs per cell).
- One and only one copy of each of the 46 chromosomes must be segregated into each daughter cell.
How is this done? Here we provide a cursory outline of DNA replication. Later in the course we will look more closely at replication, mitosis, and meiosis in the contexts of the cell division cycle, genetic inheritance, and cancer.
For now, there are three core concepts about DNA replication that you need to know:
- DNA replication is semiconservative (Fig. 8). This means that when a cell divides, the DNA duplexes in each daughter cell contain one of the parent cell’s original DNA strands (which is used as a template for polymerization, and one newly synthesized or nascent strand.
- DNA replication is semi-discontinuous (Fig. 9). This means that the nascent strand associated with one of the two strands is synthesized in short segments called Okazaki fragments that are then knitted together.
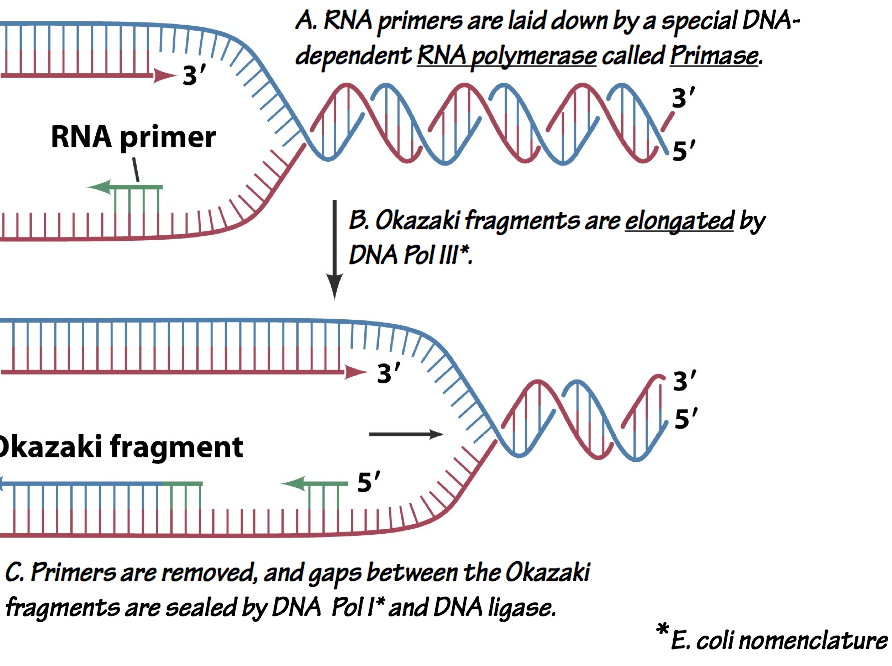
Fig. 9. Semiconservative, discontinuous replication. Each replication fork has a leading strand which is synthesized continuously, and a lagging strand which is synthesized discontinuously as a series of Okazaki Fragments. As each Okazaki fragment is completed, the preceding RNA primer is removed, and the fragments are linked, or ligated, by an enzyme called DNA ligase.
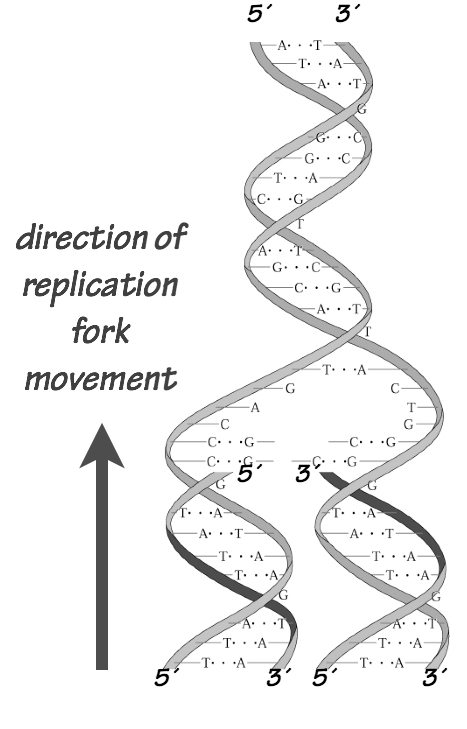
Discontinuous replication on one strand is necessary because the DNA polymerase can only add nucleotides to the 3´end of a nascent chain, but the template strands are antiparallel (Fig. 1).
An important difference between DNA and RNA polymerases is that DNA polymerases can only extend pre-existing nascent chains, while RNA polymerases can begin new chains from a single nucleotide. Thus, DNA polymerases invariably require a short RNA primer. The primer is made by a special RNA polymerase called primase (Fig. 9).
- DNA replication is bidirectional (Fig. 10). This means that at the DNA replication origin — the site where polymerization is initiated — two replication forks diverge from the
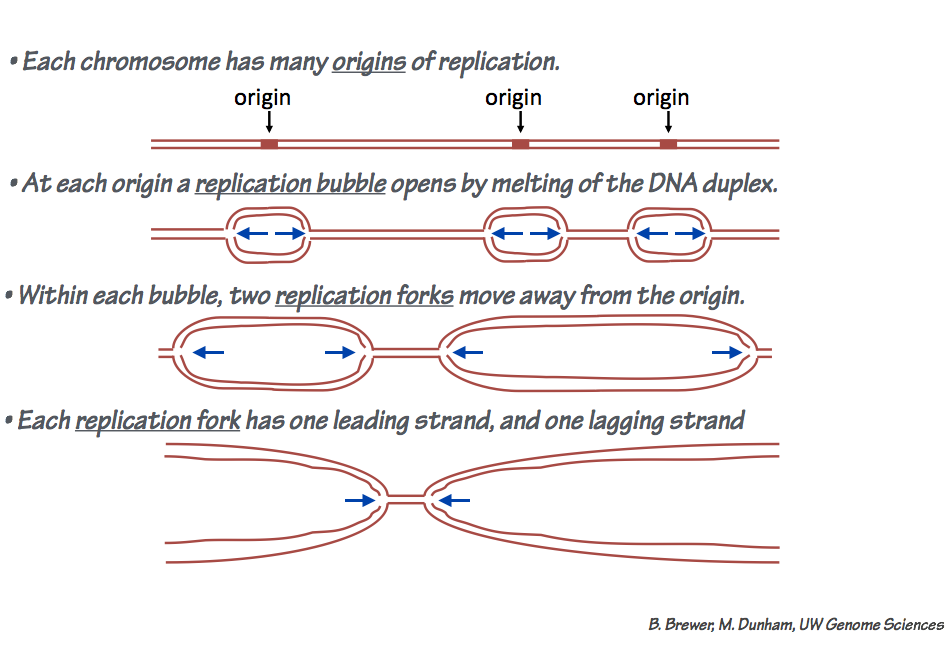
Fig. 10. Bidirectional replication of mammalian chromosomes. The DNA on each chromosome has many origins of replication. A replication bubble is opened at each origin. Each replication bubble has two replication forks that propagate away from the origin. origin. The replication machinery at each fork synthesizes one leading strand and one lagging strand (which is assembled from Okazaki fragments).
Each chromosome begins as one piece of double-stranded DNA. Replicating the very ends of the chromosomes, the telomeres, presents special problems on the lagging strand. Telomere DNA is therefore maintained by a special enzyme called telomerase.
SLO5. Summarize the structure of the peptide bond and key properties of amino acid side chains (charge, polarity/hydrophobicity, aromatic character, and reactivity) influencing polypeptide structure.
Proteins:
Proteins are the major components (by mass) of the body. Proteins have a multitude of functions: they provide structure (tensile strength; elasticity); they do mechanical work (moving chromosomes; contracting muscle); they sense the internal and external environment; they process and transmit signals in response to this information; and they carry out most of the enzymatic functions required for metabolism, for DNA replication and repair, and for gene expression. To understand the functions of proteins, we must think about their synthesis and structure.
Amino Acids & Polypeptides
A protein is made from one or more linear polypeptide chains — strings of covalently linked amino acids. The general structure of an amino acid is shown in Fig. 11. There are twenty major amino acids in eukaryotes, including humans. They differ by the side chain [R]. For the purposes of this course you do not need to memorize the structures of the amino acids. However, we will need to consider their chemical and physical properties.
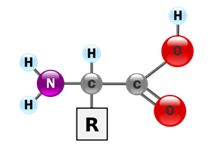
Some amino acids we can make ourselves from other chemical precursors. Others, we cannot synthesize; these must be obtained through dietary intake. We’ll consider amino acid metabolism in some detail later in the course.
Amino acid side chains are chemically diverse. Consequently, a polypeptide’s properties result from its linear sequence of amino acid residues.
- Amino acids can be hydrophilic (polar or charged), or hydrophobic (apolar; “greasy”).
- In a folded protein, hydrophilic side chains are usually exposed to the aqueous solvent. Hydrophobic side chains tend to be buried within the folded protein, so that they are shielded from the aqueous solvent.
- Amino acid side chains have diverse chemical reactivities. For example, serine threonine, and tyrosine all have terminal hydroxyl groups that can form ester bonds. Arginine and lysine contain positively charged amine groups. Cysteine contains a redox-active sulfhydryl group.
- In a polypeptide, amino acids are linked in a linear chain, head-to-tail. Linkages between amino acids are called peptide bonds (Fig. 12).
- A polypeptide backbone has a polarity: At one end, there is primary amine group. This is the amino- or N-terminus. At the other end of the backbone is a carboxylic acid group. This is the carboxy- or C-terminus. A peptide bond can be severed by hydrolysis, liberating the amine on one residue and the carboxyl group on another. The enzymes that catalyze this reaction are called proteases or peptidases.
- Almost all polypeptides synthesized by cells are synthesized one amino acid residue at a time. New residues always added to the C-terminal end of the growing chain. For this reason, we write down amino acid sequences from N-to-C.
-
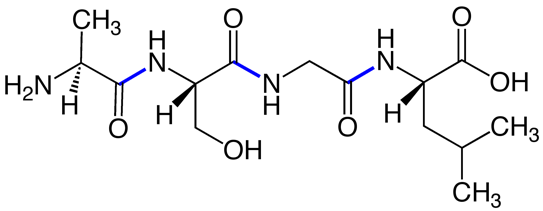
Fig. 12. Peptide bonds (blue) in a short (tetra)peptide. Source: Wikimedia The N-to-C primary sequence of a polypeptide, along with any additional covalent modifications to the polypeptide, controls how the polypeptide folds into a three- dimensional structure. This is a key point: sequence controls structure, and therefore function.
Feedback:
