Cytoskeleton, Molecular Motors, and Cell Motility
Charles Asbury
What is Cell Motility?
Session Learning Objectives
1. Compare molecular architectures of the three types of cytoskeletal filaments: microtubules, actin filaments, and intermediate filaments. Understand the structural polarity of microtubules and actin filaments, and why intermediate filaments are not polar.
2. Describe how cytoskeletal filaments provide frameworks that underlie specialized structures within cells, including the role of cell-cell and cell-matrix junctions for transmitting forces between cells. Describe how junctional defects can cause human disease.
3. Describe how a molecular motor produces movement and force via its chemo-mechanical cycle. Understand how filament polarity determines the directionality of motor movement.
4. Understand how the molecular motor dynein drives the beating motions of cilia and flagella. Describe how dysfunctional cilia can cause human disease.
5. Describe the dynamics of microtubules. Understand the importance of microtubule dynamics for mitosis, and why drugs that inhibit microtubule disassembly are used to treat cancer.
Most cells in our bodies are packed with filament networks, which are collectively known as the cytoskeleton. These filaments provide cells with rigidity and toughness to withstand mechanical perturbations. There are three types of filaments, called F-actin, microtubules, and intermediate filaments. All three are protein polymers. They are composed of thousands of identical protein subunits that assemble into much larger filamentous structures. The assembly of both F-actin and microtubules is coupled to the hydrolysis of nucleotide triphosphates (either adenosine triphosphate (ATP) or guanine triphosphate (GTP)), which provide a source of energy that allows the filaments to grow and shorten in a very dynamic fashion.
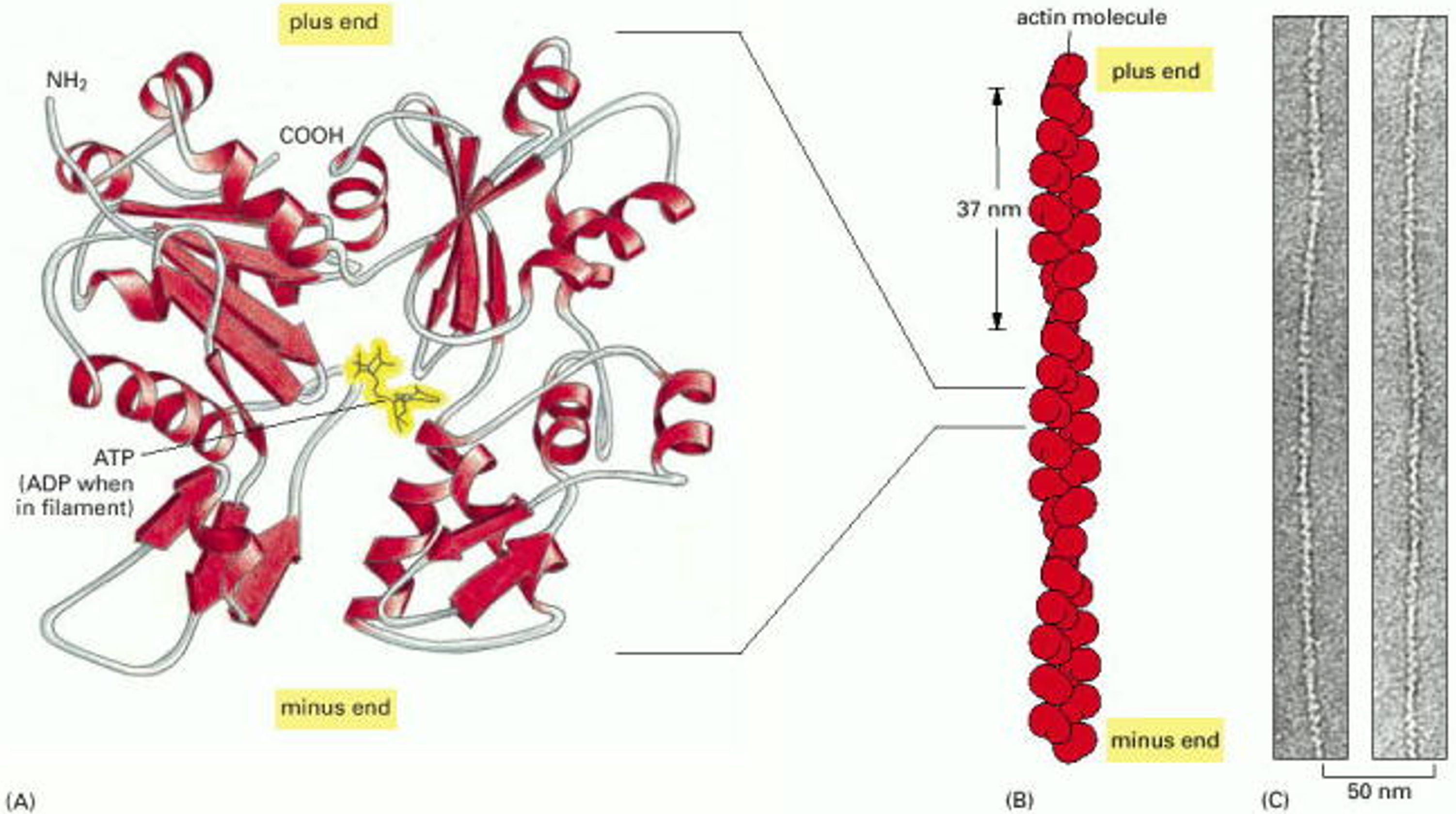
First, consider the structure of F-actin filaments (see Figure 1). These are assembled from a compact, globular protein called G-actin. (The G- stands for globular, and the F- stands for filamentous.) An individual G-actin molecule is like a very tiny LEGO block, only a few nanometers across. It has a polarity, and it has several facets where it binds to other G-actin molecules. As a consequence of the arrangement of these facets, the molecules assemble into two rows that gently spiral around one another. A typical F-actin filament may contain several hundred molecules, spanning a micrometer (10-6 m) in length. Because the individual subunits have polarity, the two ends of the filament are different. Often, the two ends are labeled ‘plus’ and ‘minus’ ends. The plus ends grow more quickly than the minus ends. (Caution: ‘plus’ does not refer to electrical charge.) The head-to-tail arrangement of subunits also gives structural polarity to filaments, allowing them to act as substrates (tracks) for directional motor transport (as discussed below in Learning Objective #3).
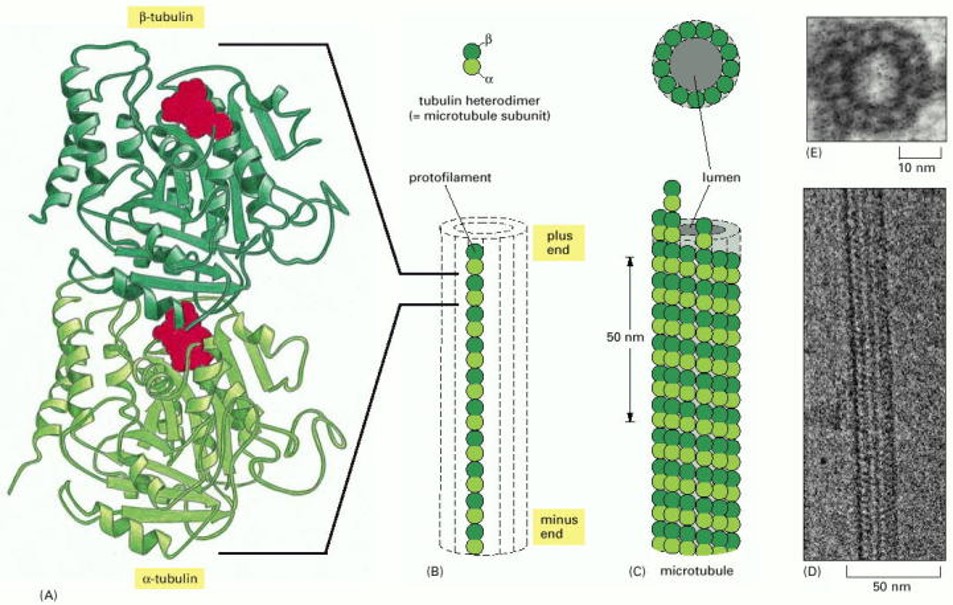
The structure of a microtubule filament (Figure 2) is conceptually similar to that of F-actin, but slightly more complicated. The elementary building block of a microtubule is a pair of globular proteins called α- and β tubulin, which associate very tightly with one another to form a heterodimer, a few nanometers in length. Each tubulin heterodimer has facets that bind other tubulin heterodimers (again like LEGO blocks), allowing them to associate in a head-to-tail fashion, and also side-to-side, to form a tiny cylindrical tube. Within the walls of the tube, heterodimers are arranged head-to-tail in longitudinal rows, called protofilaments, which run parallel to the long axis of the tube. The side-to-side bonds between heterodimers hold these protofilaments together laterally. A typical microtubule might contain several thousand heterodimers, spanning several micrometers in length. Each microtubule, like each F-actin filament, has two ends which are different, called the plus and minus ends. (Again, the structural polarity allows microtubules to act as substrates for directional motor transport, as discussed below, under Learning Objective #3. And the plus ends grow more quickly than the minus ends, a concept that will be covered in more detail under Learning Objective #5.)
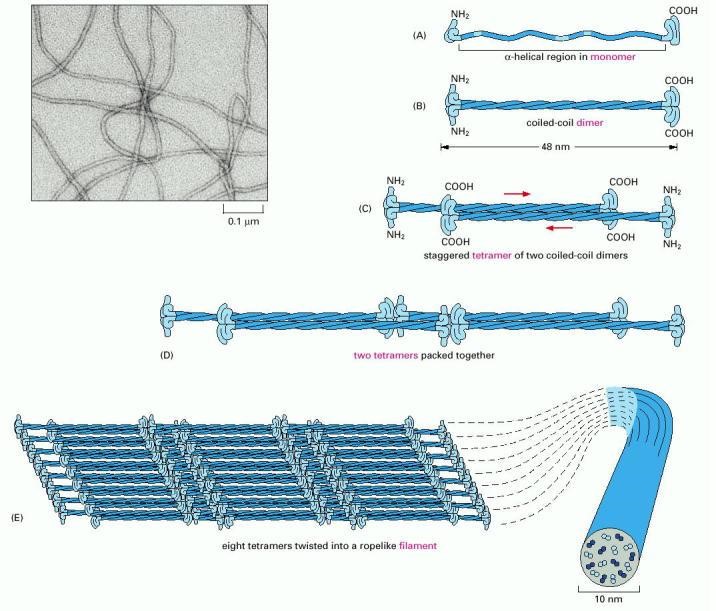
Intermediate filaments are assembled in an altogether different manner than microtubules or actin filaments. The elementary subunits do not bind nucleotides, and they are slender fibrils (rather than compact globular proteins), which pack together in a staggered arrangement (Figure 3). The subunits bind one another in an anti-parallel fashion and form filaments that lack structural polarity. As a consequence of their lack of polarity, intermediate filaments cannot provide useful tracks for motor protein-based transport. However, intermediate filaments are very important for the toughness of cells and tissues, so they can resist forces. A variety of different intermediate filaments exist, including keratins (found in skin and hair), desmins (found in muscle), and neurofilaments (brain and other neurons). The diversity of intermediate filaments makes them useful for identification of different types of cells.
Simple exercises to check what you recall
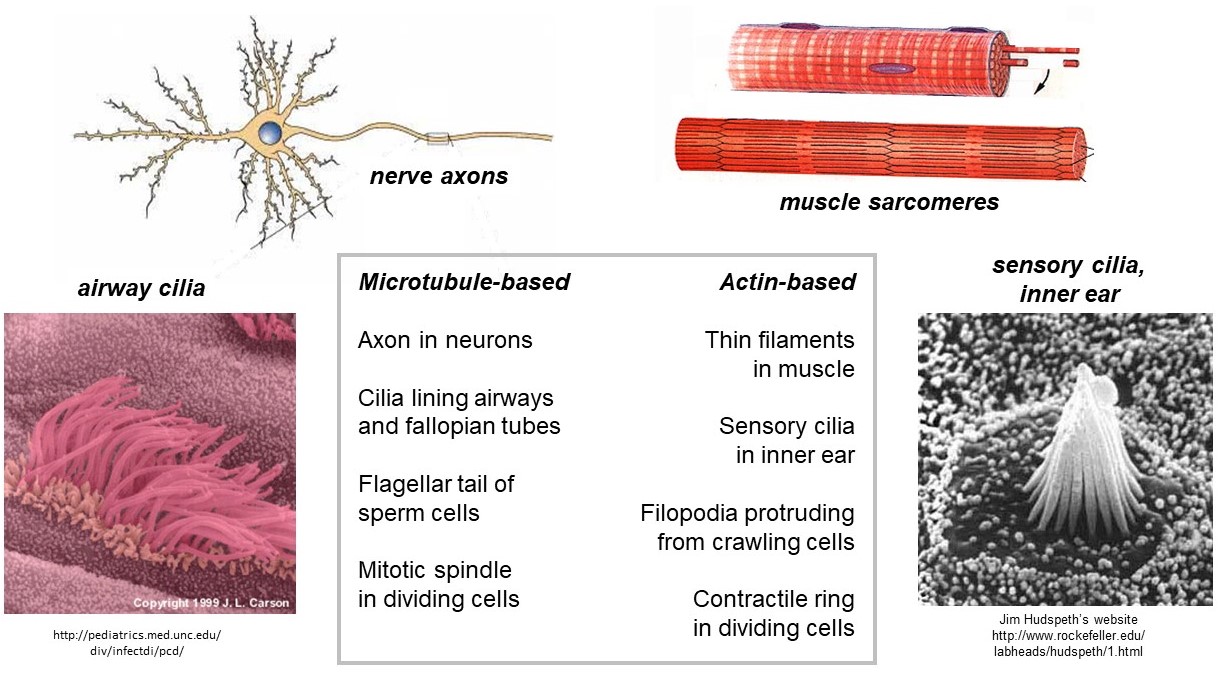
Cytoskeletal filaments provide the framework for various specialized structures in cells (Figure 4). The filaments in these structures often have identical polarity, with plus ends pointing in the same direction. One such structure is the muscle sarcomere. The thin filaments within sarcomeres are composed primarily of F-actin filaments with their plus ends all embedded in the z-bands (z-disks) on the left and right sides, and their minus ends pointing toward the middle (the m-line). Other examples of specialized structures that contain polarized F-actin are (i) the sensory cilia in the inner ear, responsible for hearing, (ii) the filopodia that protrude from crawling cells (e.g., of our immune systems), and (iii) the contractile ring which drives fission of dividing cells. Polarized arrays of microtubules are found in (a) the axons of nerve cells, (b) the cilia lining our airways, (c) the flagellar tail of sperm cells, and (d) the mitotic spindle that drives chromosome separation in dividing cells.
Cytoskeletal junctions allow force transmission between neighboring cells
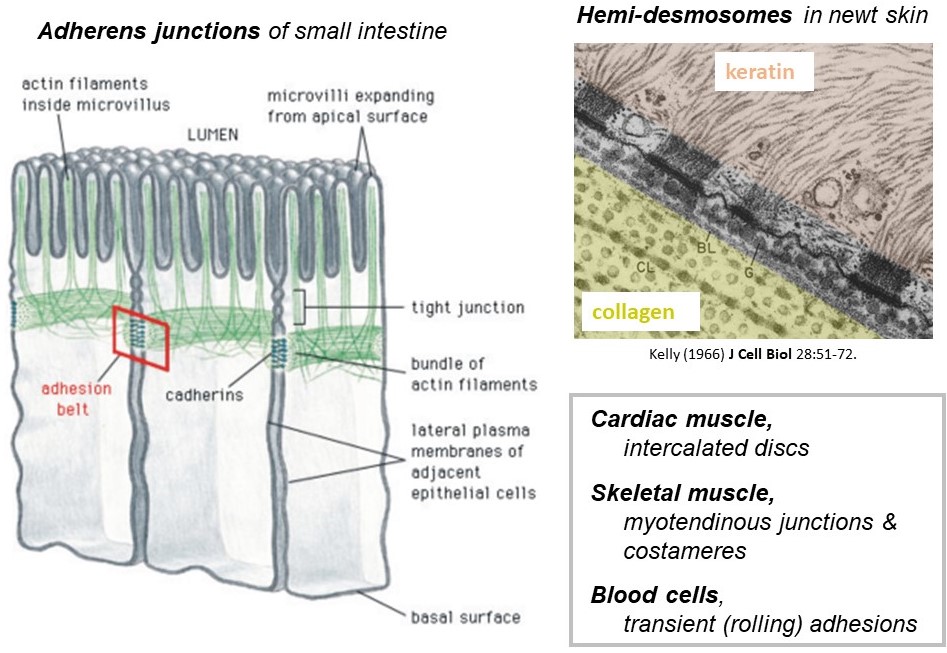
The cytoskeletal filaments described above are found exclusively inside cells and, as already mentioned, they provide toughness that allows the cells to withstand mechanical forces. The forces carried by cytoskeletal filaments are transmitted from one cell to a neighboring cell, or to the extracellular matrix, by specialized junctions (Figure 5). The junctions are collections of membrane-spanning proteins that connect to cytoskeletal filaments inside the cell, to identical junctional proteins in the plasma membrane of a neighboring cell, or to extracellular matrix proteins. There are many different types of junctions found in different cellular contexts. Some examples are the adherens junctions that connect F-actin filaments in neighboring epithelial cells lining the lumen of your small intestine, and the hemi-desmosomes that connect keratin intermediate filaments inside your skin cells to extracellular matrix fibers such as laminin on the outside. The intercalated discs that connect cardiac muscle cells to one another, and the myotendinous junctions that connect skeletal muscles to tendons are also examples of cytoskeletal junctions.
Simple exercises to check what you recall
Axonal transport: single motors hauling vesicles
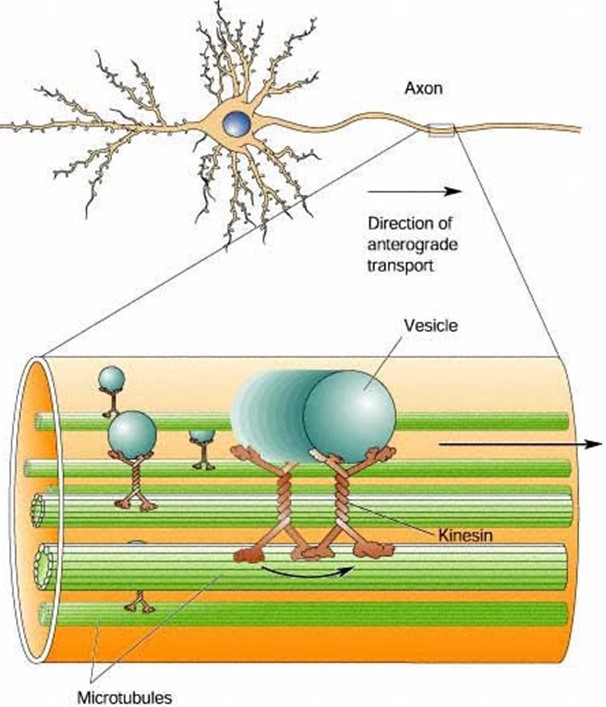
Because of their highly elongated shape, neurons rely heavily on intracellular transport. Smaller cells can rely on diffusion to deliver proteins, metabolites, second messengers, and so forth, from their sites of synthesis to their sites of action. But diffusion is far too slow in long, slender neurons. Consider, for example, the extraordinary length of individual motor neurons innervating your toe. These cells have axons that project all the way from the base of your spine to your toe, about 1 meter away. Even the smallest, fastest diffusing metabolites would take years to diffuse from where they are made, in the cell body (the soma, in your spine) to where they are needed, at the end of the axon (neuromuscular junction, in your toe). These extremely long cells must rely on active transport to carry cargo down their axons.
What defines a molecular motor?
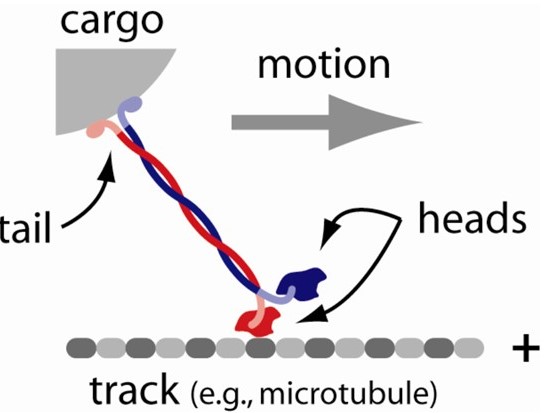
Molecular motors are amazing enzymes that convert chemical energy, usually in the form of adenosine triphosphate (ATP), into mechanical work. The myosins that power muscle contraction are the most famous molecular motors. But hundreds of other motors are found in human cells. Luckily, most of them operate by the same general principles (Figure 7).
Generally, a molecular motor…
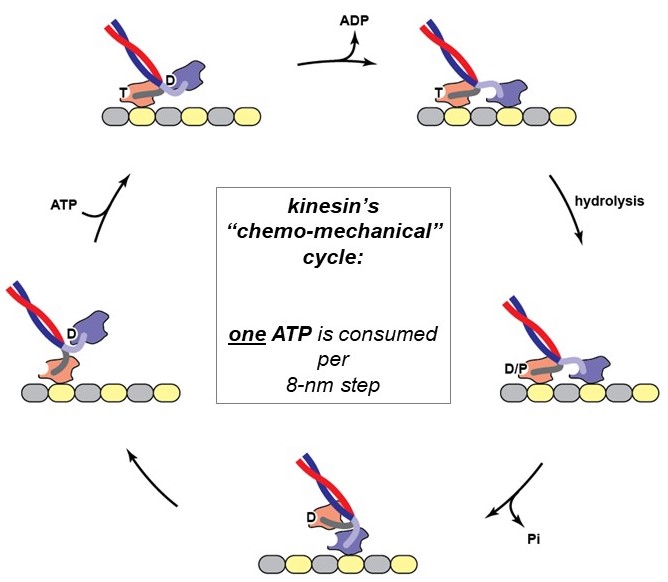
The action of a motor protein is strikingly sophisticated, and similar in many ways to human-made machines: The mechanical cycle of a motor protein’s heads (attach, stroke, detach, reset) is tightly coupled to a biochemical cycle that includes binding of an ATP (fuel) molecule, hydrolysis (breakage) of the high-energy gamma-phosphate bond of the ATP to produce ADP and phosphate, and then release of these reaction products. Because chemical and mechanical activities are coupled together, this process is called the chemo-mechanical cycle of the motor. The chemo-mechanical cycle of kinesin is diagrammed at right (Figure 8) and animated in the 2-minute video below (includes narration).
The movement produced by molecular motors can be observed directly in a microscope using an in vitro motility assay. For example, kinesin motors attached to a glass surface will drive ATP-dependent gliding of microtubule filaments, as seen in this 14-second video (this one has no sound):
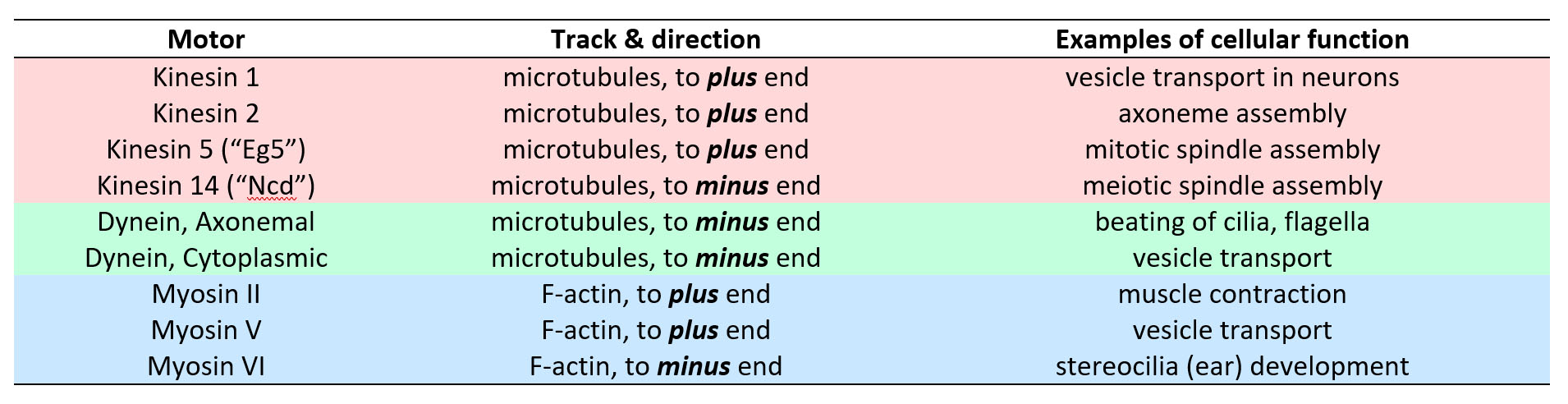
Most motors that participate in cell motility fall into three families, the kinesin, dynein and myosin families (see Table 1). These families are defined by the detailed molecular architecture of the motors, which is beyond the scope of this class. However, a few general points are worth remembering. First, all motors from the myosin family move along F-actin filaments, and all motors from the kinesin and dynein families move along microtubules. Second, motors with opposite directionality can be found within the families. That is, there are some kinesin motors that move toward the microtubule plus end, and some that move toward the minus end. The same goes for myosins, and probably also for dyneins.
Some motors work in groups, others work alone
Simple exercises to check what you recall
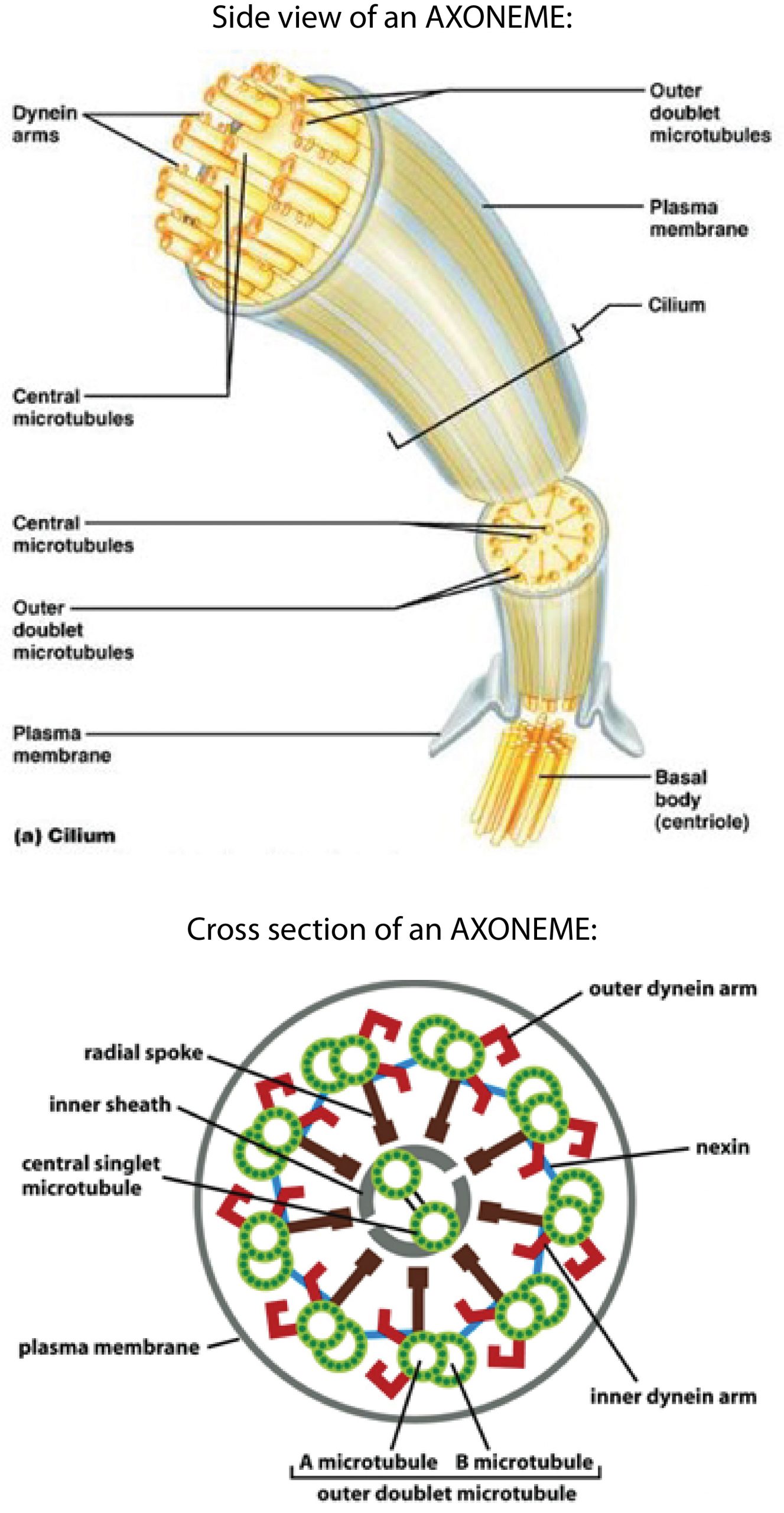
The whip-like movements of cilia and flagella are medically relevant examples of cell motility driven by molecular motors. Motile cilia are important for clearing mucus (and embedded pathogens) from our airways, for moving eggs through the fallopian tube to the uterus, and for circulating cerebrospinal fluid through the brain and spinal cord. Flagella, which are morphologically and functionally identical to cilia, drive sperm movement. Both cilia and flagella are driven by large collections of dynein motors. We will focus on cilia; but the same principles apply to flagella and cilia.
How ciliary movement is generated
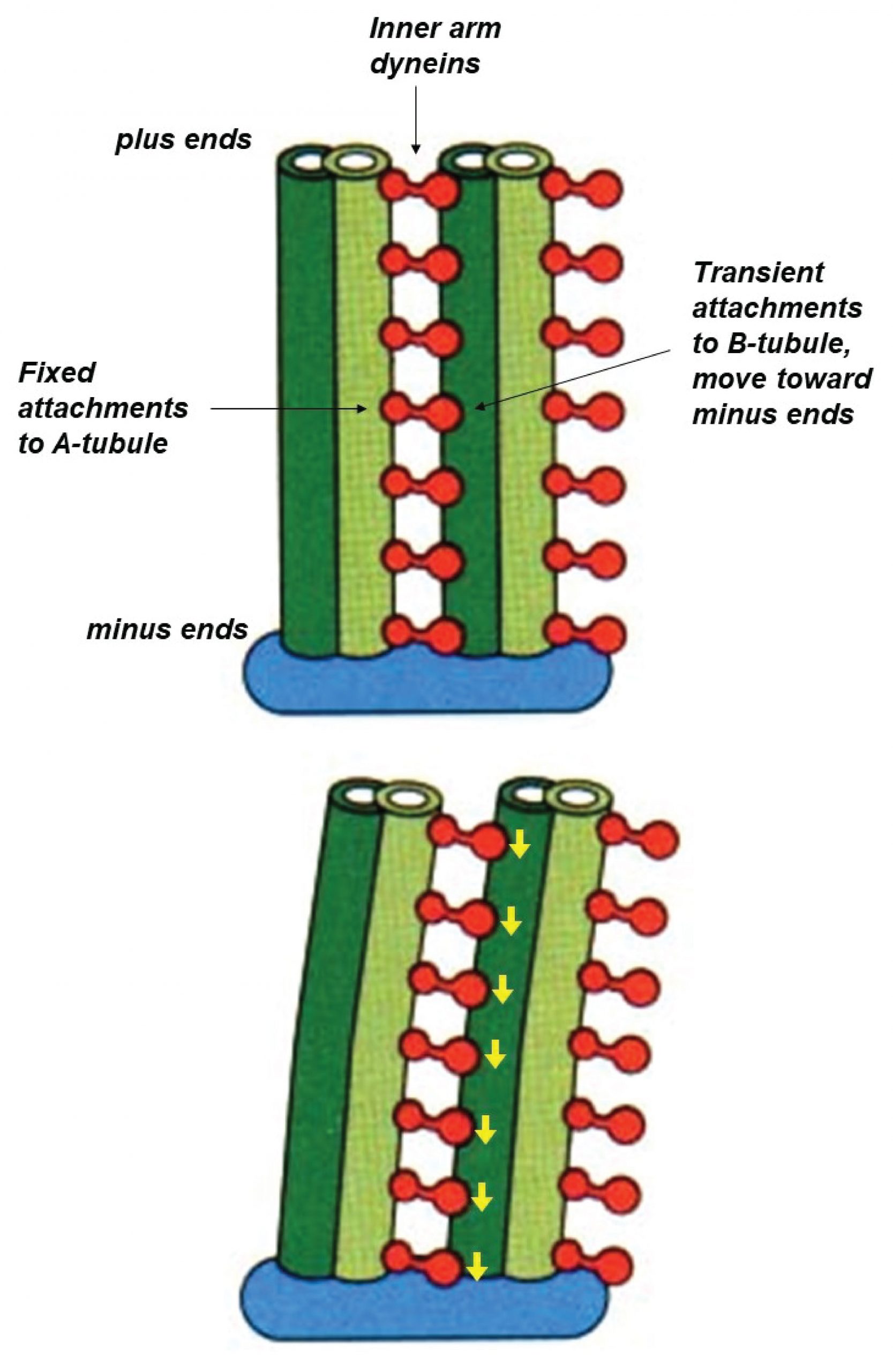
Each ciliary dynein motor is permanently fastened by its tail to one microtubule. Its head can reach across to a neighboring microtubule, making transient attachments that move toward the minus end at the base of the axoneme. These movements tend to drive the filaments to slide past one another (similar to the relative sliding of thick and thin filaments in muscle). However, in the axoneme the filaments are anchored together at their base. This anchoring resists the sliding movement and, as a consequence, the sliding is converted into bending (Figure 10).
Non-motile cilia often serve as signaling ‘antennas’
Dysfunction of cilia causes diseases known as ‘ciliopathies’
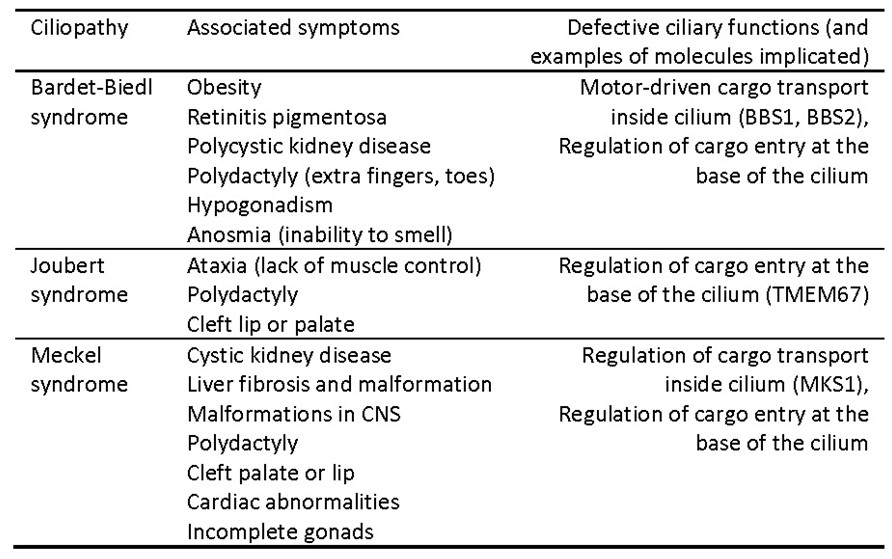
The wide variety of problems that can result from impaired cilia is a testament to the vital importance of cilia for human health. Defects in non-motile sensory or primary cilia can cause kidney disease, blindness, obesity, abnormal heart formation, polydactyly (extra fingers or toes), cleft palate, and other developmental malformations (see Table 2). Defects in the movement of motile cilia can cause severe mucous buildup in the lungs, and infertility. A host of rare, congenital syndromes, each associated with a constellation of symptoms that naively seem unrelated, are now recognized as ciliopathies, caused by various defects in ciliary function. Three example ciliopathies, outlined in Table 2, are caused by impairment of molecules that control entry of cargoes into the cilium or regulate intra-flagellar transport within the cilium. The very broad range of defects can be explained by the central role that intra-flagellar transport plays in delivering many cargoes required for ciliary signaling in many different tissues.
Simple exercises to check what you recall
The emphasis above on motor-based movements might give the impression that cytoskeletal filaments play a static role in cell motility, acting solely as tracks along which molecular motors move. While this is true in some contexts (e.g., nerve axons, motile cilia, and muscle sarcomeres), in other contexts, the filaments play a much more dynamic role. Below is a brief (7-second) movie, where you can see dynamic F-actin (tagged with a fluorescent marker) at the leading edge of a cell that is crawling upward in the image. The leading edge of the cell is pushed upward by the rapid growth of the F-actin filaments, which are simultaneously flowing backwards (this movie has no sound):
In dividing cells, bundles of microtubules constantly grow and shorten, pushing and pulling the chromosomes back-and-forth. To understand how filament dynamics can contribute to cell motility, it is important to understand the dynamics exhibited by the filaments alone. We will focus on microtubules.
The ‘dynamic instability’ of microtubule filaments
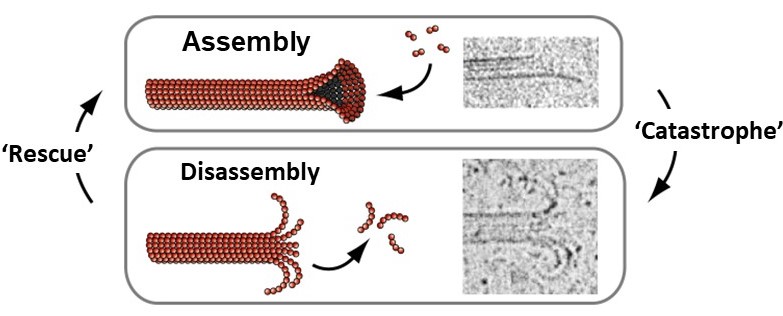
Microtubules spontaneously grow and shorten by addition and loss of tubulin subunits from their tips (Figure 11). They also exhibit a behavior called dynamic instability, that can be summarized as follows: In solutions containing free tubulin and GTP, growing and shortening microtubules coexist. When individual filaments are followed over time, they randomly switch back-and-forth between growth and shortening, exhibiting large fluctuations in length, as seen in this 32-second video (includes narration):
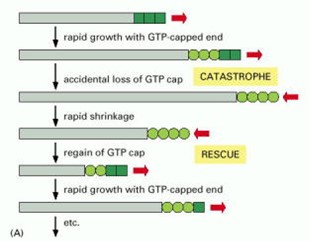
Each of the tubulin subunits that makes up a microtubule is an enzyme, containing a pocket that binds guanine nucleotides (see Figure 2) and catalyzes nucleotide hydrolysis. The dynamic instability of microtubule filaments is powered by this hydrolysis of GTP (Figure 12). Under physiological conditions, only GTP-containing tubulin is competent for assembly onto growing filament tips. GDP-containing tubulin cannot assemble. Thus, newly-added subunits at growing microtubule tips contain GTP (forming a ‘cap’ of GTP-tubulin). As more growth occurs these subunits become buried deeper in the filament wall, where they are stimulated to hydrolyze their GTP, releasing phosphate but retaining GDP. Hydrolysis causes a conformational change in the tubulin that weakens side-to-side interactions between neighboring subunits in the microtubule lattice, thereby destabilizing the GDP-containing portion of the filament. Despite this destabilization, the GDP lattice will remain intact and the filament will continue to grow, provided that a GTP cap exists at the tip. If the GTP cap is lost, however, the GDP lattice quickly disassembles. This explains why microtubules can suddenly switch from slow growth to rapid shortening, a transition colorfully named a ‘catastrophe’. Shortening microtubules can also suddenly resume growth, a transition known as a ‘rescue’, which presumably occurs when the GTP cap spontaneously re-forms.
F-actin dynamics and treadmilling
F-actin filaments exhibit a treadmilling behavior, which differs from the dynamic instability of microtubules but shares a dependence on nucleotide hydrolysis. G-actin is an enzyme with a pocket for binding and hydrolyzing ATP (see Figure 2). Its assembly onto filaments occurs predominantly by addition of ATP-containing monomers. Incorporation into a filament stimulates the monomers to hydrolyze ATP and release phosphate, so the filaments retain only ADP. Because the assembly and disassembly processes are not chemical reversals of one another, it is possible for the two ends to behave quite differently: For example, one end can be assembling while the other is disassembling. This is called treadmilling.
Common features of dynamic protein polymers
The following features are shared by the dynamic protein polymers, F-actin and microtubules:
(i) Filaments spontaneously grow by addition and loss of subunits from their tips.
(ii) Each subunit has a binding pocket for nucleotide (G-actin binds ATP, tubulin binds GTP).
(iii) Assembly occurs by addition of NTP-bound monomers to tips.
(iv) Assembly into filament promotes NTP hydrolysis. Thus, apart from the freshly-assembled cap, filaments retain only NDP-bound subunits.
(v) Disassembly occurs primarily by loss of NDP-bound forms from tips.
Dynamic filaments drive many cellular movements
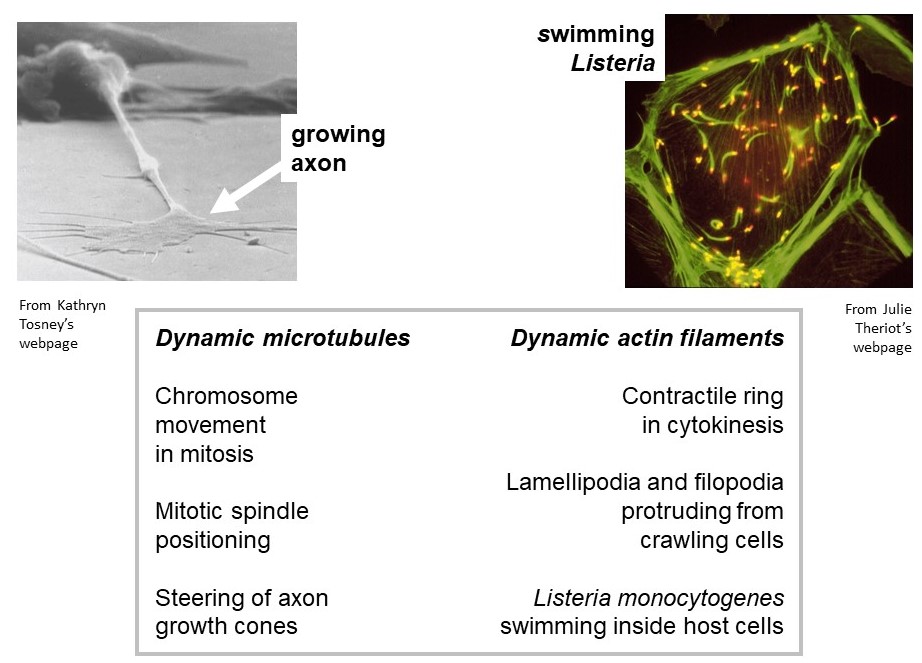
Actively growing/shortening actin and microtubule filaments drive many types of non-muscle cell motility. Examples of cell motility that require microtubule growth and shortening are (a) positioning of the mitotic spindle within a dividing cell, and (b) steering of axon growth cones during neural development. Examples of motility driven by dynamic actin filaments are (i) the protrusion of lamellipodia and filopodia from the leading edge of crawling cells, and (ii) the swimming movement of the infectious bacterium, Listeria monocytogenes, in the cytoplasm of a host cell. (Listeria infection is the cause of listeriosis, a type of food poisoning. To see listeria swimming, check out: https://www.youtube.com/watch?v=sF4BeU60yT8.)
Mitosis: dynamic microtubules move chromosomes
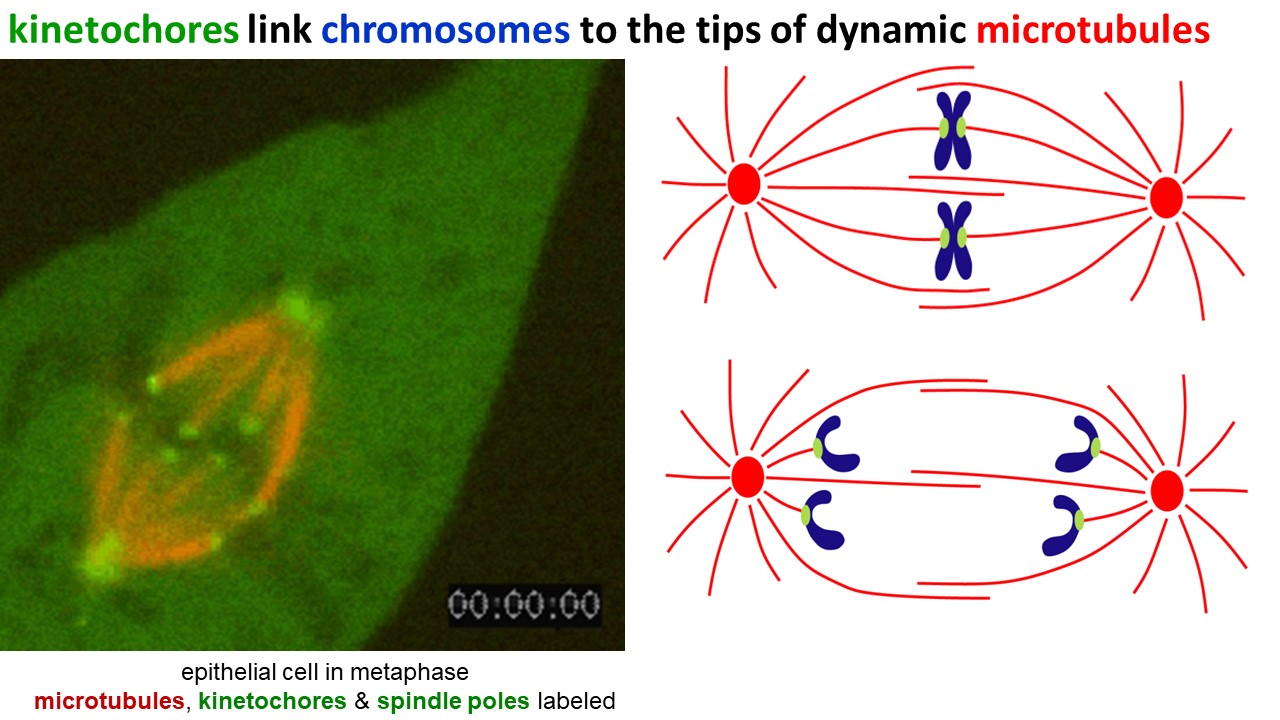
Dynamic filaments are vital for mitosis, the process by which duplicated chromosomes are segregated during cell division. In cells preparing to divide, microtubules assemble into a bipolar array called the mitotic spindle (Figure 14). Spindle microtubules are highly dynamic – constantly growing and shortening. Their tips capture the duplicated chromosome pairs. While the chromosomes are captured, the growing and shortening microtubules push and pull them to the equator of the spindle, and then separate each pair so that when cell cleavage occurs, each daughter cell gets a complete copy of the genome.
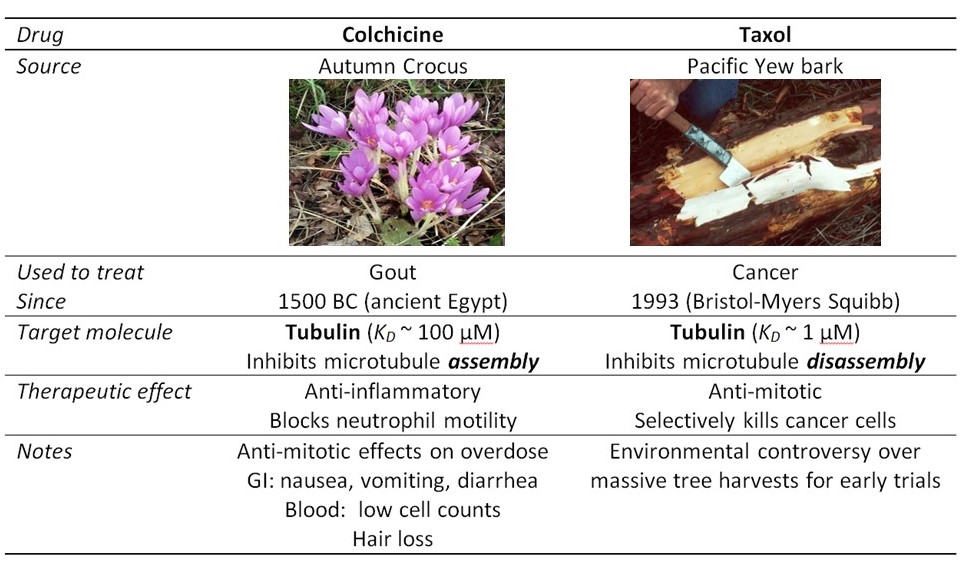
Accurate mitosis is essential for the development of a human baby, composed of trillions of (1012) cells, from a single embryonic cell. Mitosis is also critical for maintenance of healthy tissues. Many tissues in our body contain populations of stem cells, which continually divide to replenish dead cells (e.g., in our blood stream, skin, and intestines). Mitosis also has a role in cancer: Abnormal mitosis, where the daughter cells inherit broken, extra or missing copies of a chromosome, contributes to the progression of certain types of cancer. Moreover, drugs that block the activity of the mitotic machinery are often used as chemotherapeutics (Table 3). Such anti-mitotic drugs act by inhibiting the rapid, out-of-control cell division of tumor cells. They also block division of healthy cells, and therefore cause other side-effects that limit the dose a patient can withstand.
Two excellent, short videos of mitosis
Mitosis in a lily plant cell, one minute (includes narration):
Mitosis in a newt lung cell, one minute (includes narration):
Simple exercises to check what you recall
Extra Q&A from past students that might be helpful
Hi Dr. Asbury,
I have a question regarding Monday’s IRAT. Question number 5 asked “How do the nucleotides ATP and GTP participate in non-muscle motility” and one of the options was “ATP is an energy source driving actin filament ‘treadmilling’, where one end assembles while the other simultaneously dissembles”. I don’t remember that being talked about in the syllabus and I am wondering if you can explain what this means, given it was a true statement? If maybe I overlooked it, can you direct me to which page this is talked about on?
Thanks so much for contacting me, and for your question about treadmilling of F-actin. I think there are perhaps three questions underlying your email, and I will try to address them all here:
First, what do I mean by “treadmilling”? The individual actin subunits can each bind and hydrolyze ATP — in other words, each one is an ATP-splitting enzyme, and like the other ATP-splitting enzymes discussed in our course (myosin, kinesin, dynein, SERCA…), they harness energy from the splitting reaction in order to drive dynamic processes. The ATP-splitting that occurs within F-actin drives treadmilling, which is when G-actin subunits are being added to one end of a filament while they are simultaneously being lost from the other end. This happens because the subunits must contain ATP in order to bind onto the end of a filament, and because after they are bound into the filament, they are stimulated to split their ATP and release the phosphate, (retaining only ADP). The splitting reaction takes a little time, so the newly grown filament tip is composed of freshly added ATP-containing subunits, bound on top of ADP-containing subunits that have resided inside the filament for more time. We say the filament is “capped” with ATP-containing actin. The ADP-containing actin does not bind as tightly to its neighbors in the filament, so if it happens to be exposed at the opposite end of a filament, it typically falls off. The overall result is that you can have a growing end where ATP-containing actin is getting added, while simultaneously at the opposite end of the filament there is ADP-actin falling off. We call this “treadmilling” because it is like a treadmill, with the actin subunits moving through the filament from one end to the other. This movement is powered by ATP hydrolysis.
Your second question is whether and where this “treadmilling” was described in the syllabus. And underlying that question, I think you are probably wondering, will I be expected to know all this for my upcoming quiz? This Pressbook course pack only briefly mentions actin dynamics. It mentions briefly how the polarity of F-actin allows the two ends of the filaments (the “plus” and “minus” ends) to grow at different rates. It also mentions that dynamic F-actin pushes the leading edges of crawling cells forward, and that it drives movement of Lysteria, an infectious bacterium, inside host cells. I chose to focus this Pressbook course pack more on the very similar dynamics of microtubules. For brevity, our coverage of F-actin dynamics is only very limited. This is also true during my in-class session, and in the pre-class movies that Dr. Jay Gatlin (at U Wyoming) prepared.
Thus, my expectation is for you to understand tubulin dynamics in a little bit more detail than actin dynamics, and I do not expect you to understand the details of actin treadmilling specifically. The two most important general concepts related to filament dynamics are that (1) energy supplied by the hydrolysis of nucleotides — GTP for tubulin, ATP for actin — is harnessed by these protein polymers to make them assemble and disassemble very dynamically in some situations, and that (2) these nucleotide hydrolysis-powered dynamics can generate pushing and pulling forces, for example to move the front of a crawling cell forward (an actin-driven process) and to separate chromosomes during cell division (a microtubule-driven process).
Feedback:
a network of protein filaments in the cytoplasm of living cells, supporting their shape
the polymerized, filamentous form of actin (the F- stands for filamentous)
the polymerized, filamentous form of α- and β tubulin
one of three types of cytoskeletal filaments
the unpolymerized, non-filamentous form of actin (the G- stands for globular)
the head-to-tail arrangement of subunits within an F-actin or microtubule filament; polarity is a structural property of F-actin and microtubules
the unpolymerized, non-filamentous subunits of microtubules; a heterodimer of α- and β tubulin represents one subunit of a microtubule
a type of intermediate filament found in skin, hair, and nails
a type of intermediate filament protein found in muscle cells
a type of intermediate filament protein found in the brain and other neurons
the smallest functional unit of a myofibril in striated muscle
cell-cell adhesion complexes whose cytoplasmic faces link to the F-actin cytoskeleton; sometimes called "zonula adherens", "fascia adherens" (e.g., in cardiac cells), or "belt desmosomes"
cell-matrix adhesion complexes whose cytoplasmic faces link to keratin intermediate filaments inside a cell and whose extracellular faces link to fibers outside such as laminin
specialized adhesion structures that connect adjacent cardiac muscle cells
specialized cell adhesion structures that connect tendon to muscle
movement away from the cell body, or 'soma' of a neuron
movement toward the cell body, or 'soma' of a neuron
one of three families of ATP-powered motor proteins; kinesins move along microtubule filaments
one of three families of ATP-powered motor proteins; dyneins move along microtubules
the cyclical action of a motor protein, through which a sequence of biochemical transitions associated with nucleotide hydrolysis are coupled to a sequence of mechanical transitions associated with movement
a method for directly observing the motion generated by purified molecular motors
one of three families of ATP-powered motor proteins; myosins move along F-actin
the ability of a molecular motor to generate many stepwise movements along its track without detaching
a microtubule- and dynein-based machinery inside cilia and flagella that drives their beating movements
a single, non-motile cilium found on almost every human cell
the movement of cargoes up and down a cilium driven by kinesin and dynein motors
diseases caused by defects in the function of cilia
the process by which microtubules switch suddenly between periods of growth and periods of shortening
infection caused by the germ Listeria monocytogenes, usually occurring after eating contaminated food
the segregation of duplicated chromosomes to opposite sides of a dividing cell before it pinches into two daughter cells
the microtubule-based machinery that organizes and separates duplicated chromosomes during cell division
drugs that block cell division by interfering with the function of the mitotic spindle

